DDBJ fruther upgraded ARSA (All-round Retrieval of Sequence and Annotation) is the high-speed keyword search system provided by DDBJ.
The features of ARSA are as follows:
- ARSA provides one-stop query to 24 life science databases (30 million entries) including DDBJ International Nucleotide Sequence Database.
- ARSA will return a quick response to any query regardless of the complexity of the query, the number of hits to the query and the target database(s)
- In the case of DDBJ, you can define search conditions in details by use of any combination of Features/Qualifiers of the DDBJ flat file.
- Application Programming Interface (API) is prepared so that you can call some functions of ARSA from your programs such as Java, in your computer.
DDBJ added the following new functions this time.
- Addition of the KEGG/PATHWAY database to the searchable databases
KEGG/PATHWAY database was newly added. You can view the PATHWAY maps by clicking the PATHWAY IDs in the result lists.
- Specification of detailed search conditions in the all databases, as well as DDBJ
The detailed search conditions can be specified in the all databases (When you use this function, select the database in the top page, then click the "Cross Search" button).
- Cross Search of multiple databases using the common search conditions
When you select the multiple databases, you can use the above function by using the common search conditions (Check the multiple databases you would like to search in the top page, then click the "Cross Search" button).
Other improvements:
- "&(AND)" and/or "|(OR)" can be entered into the search boxes directly
(e.g. see the "Help" page)
- The restriction for using the following words in the refine search was removed.
(In the new ARSA, " ' ", " ( ", " ) ", " { ", " } " can be used as the search keywords)
-
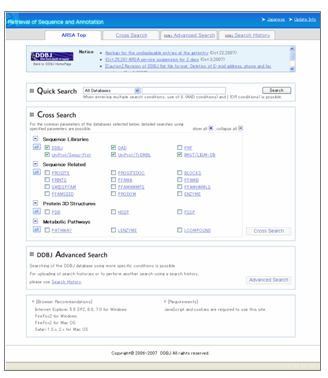
ARSA New Top Page
|
Hit Count List Page
|
ARSA will continue to be enhanced, if you make comments. So, please visit ARSA and click "Your Comment" to help us to improve ARSA to help you.

