New BioProject, BioSample and DRA submission system were released at 8th, April (Wed).
This system enhancement was carried out by funding from MEXT Genome Science.
Tutorial video.[youtube id="Hg4h8ytbIV4" title="Submission to BioProject, BioSample and DRA"]
The new system is able to reference and submit BioProject, BioSample and DRA submissions which had been registered in the former system.
BioProject, BioSample and DRA data can be submitted at once
Before submission to DRA, BioProject and BioSample accession numbers were required.In the new system, BioProject, BioSample and DRA data can be submitted at once, so submitters need not to wait for BioProject and BioSample accession numbers.
Experiment and Run which are same number of BioSample, are automatically created.Submitter and hold action in the DRA submission are copied to "SUBMITTER" of BioProject and BioSample.
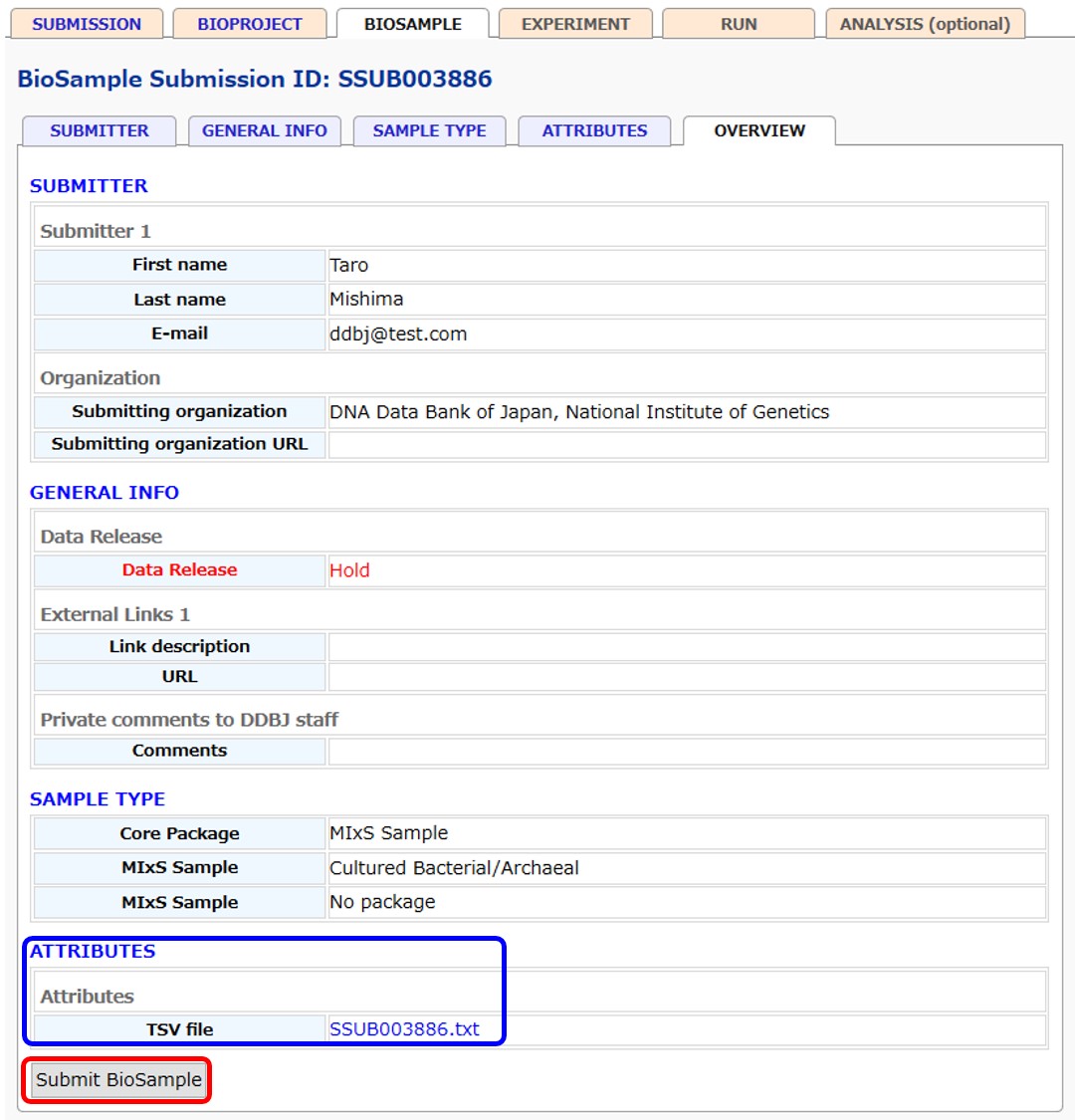
Registered BioSample attributes text file is available in D-way
Users can download the registered BioSample attributes text file in D-way (blue box).Updates will be reflected to the file.
Reference sequence of bam can be specified by accession number
Reference sequence in single fasta file was required to submit alignment data in bam.
In the new system, if references are found in ftp://ftp-trace.ncbi.nlm.nih.gov/sra/refseq/,references can be specified by their accession numbers.
In the DRA submission tool, "BioSample Used" was moved
In the DRA submission tool, "BioSample Used" was moved from rightmost to just right to "Experiment Alias",which enables users to easily understand relationship between "Experiment" and "referencing BioSample".
Fastq from capillary sequencing platforms can be submitted to DRA
In the DRA submission tool, capillary sequencing platforms can be selected.Fastq from these platforms can be submitted to DRA.To submit sequence chromatgrams, please continue to submit data to DDBJ Trace Archive.
- AB 3730xL Genetic Analyzer
- AB 3730 Genetic Analyzer
- AB 3500xL Genetic Analyzer
- AB 3500 Genetic Analyzer
- AB 3130xL Genetic Analyzer
- AB 3130 Genetic Analyzer
- AB 310 Genetic Analyzer
Sequencing instruments were added
In the DRA submission tool, following Illumina and Oxford nanopore sequencers were added.
- HiSeq X Ten
- NextSeq 500
- MinION
- GridION
PacBio hdf5 file can be submitted
PacBio hdf5 file can be submitted.Select "PacBio_HDF5" for Run File Type.
"Illumina_native", "Illumina_native_qseq" and "Helicos_native" are removed from Run filetype
"Illumina_native", "Illumina_native_qseq" and "Helicos_native" are removed from Run filetype. These types can be specified in XML.
Other changes
-
BioProjectand BioSample Submitting organizations are pre-entered with "Lab/Group", "Department (2)", "Department (1)", "Organization" of D-way account.Text in the submitting organization can be editted. - BioSample "Private comments to DDBJ staff" was moved from "COMMENTS" tab to "GENERAL INFO". "COMMENTS" tab was removed.