WABI BLAST (Tentative closing)
Overview
WABI provides web APIs for using BLAST.
- A search job is registered in a queue by submitting a
BLAST search criteria. A Request
ID is returned as response data.
Once a search job is registered in the queue, the state of the job transitions from “Waiting” ==> “Executing” ==> “Completed”.
Once the status of a job moves to the “Completed” state, the corresponding search results will be available for viewing. Results are retained on the server and can be viewed for up to 7 days after the search has been executed.
(Refer to “Search result retention period” 「Request ID and BLAST result」 ) - Returns the status of the BLAST search for a specified Request ID.
- Returns the BLAST search criteria for a specified Request ID.
- Returns the BLAST search result for a specified Request ID.
Searching WABI BLAST
BLAST search job
A BLAST search job is first registered in the queue of jobs awaiting execution. The job is then executed as soon as computing resources become free.
See Also: Search job status
Search Job Status
WABI search jobs transition through the following status.
| Job Status | Explanation |
|---|---|
| waiting | The job has been added to the queue, but has not yet commenced execution. |
| running | The job is being executed. |
| finished | The job has completed execution. |
| not-found | The job with the specified Request ID does not exist. |
Example response values when querying job status:
1 request-ID: wabi_blast_1111-1111-1111-11-111-111111
2 status: finished
3 current-time: 2013-01-01 12:34:56
4 system-info:
5 stdout
1 {
2 "request-ID": "wabi_blast_1111-1111-1111-11-111-111111",
3 "status": "finished",
4 "current-time": "2013-01-01 12:34:56",
5 "system-info": "nstdout"
6 }
1 <?xml version="1.0" ?>
2 <!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
3 <result>
4 <request-ID>wabi_blast_1111-1111-1111-11-111-111111</request-ID>
5 <status>finished</status>
6 <current-time>2013-01-01 12:34:56</current-time>
7 <system-info>stdout</system-info>
8 </result>
1 {
2 "error-message": "Unexpected error (status == null)",
3 "requestId": "wabi_blast_1111-1111-1111-11-111-111111",
4 "jobId": "123456",
5 "month": "01",
6 "day": "01",
7 "hour": "12",
8 "min": "34",
9 "sec": "56",
10 "millisec": "789",
11 "randomId": "987654"
12 }
BLAST search criteria
Example of parameters that are passed on when a BLAST job is submitted using WABI.
Example:
1 {
2 "address": "",
3 "database": "hum",
4 "datasets": "ddbjall",
5 "format": "json",
6 "parameters": " -v 100 -b 100 -e 10 -F F -W 11",
7 "program": "blastn",
8 "querySequence": ">my query sequence 1nCACCCTCTCTTCACTGGAAAGGACACCATGAGCACGGAAAGCATGATCCAGGACGTGGAAnGCTGGCCGAGGAGGCGCTCCCCAGGAAGACAGCAGGGCCCCAGGGCTCCAGGCGGTGCTGnGTTCCTCAGCCTCTTCTCCTTCCTGCTCGTGGCAGGCGCCGCCACn",
9 "result": "www"
10 }
BLAST search result
The following is an example of the BLAST search output. This is the same as the file generated using the blastall command, when executed using the “-o” command line option.
Example:
1 BLASTN 2.2.25 [Feb-01-2011]
2
3 Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
4 Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
5 "Gapped BLAST and PSI-BLAST: a new generation of protein database search
6 programs", Nucleic Acids Res. 25:3389-3402.
7
8 Query= AB000095|AB000095.1 Homo sapiens mRNA for hepatocyte growth
9 factor activator inhibitor, complete cds.
10 (1740 letters)
11
12 Database: hum
13 572,091 sequences; 5,019,832,159 total letters
14
15 Searching..................................................done
16
17 Score E
18 Sequences producing significant alignments: (bits) Value
19
20 AB000095|AB000095.1 Homo sapiens mRNA for hepatocyte growth fact... 3449 0.0
21 BC018702|BC018702.1 Homo sapiens serine peptidase inhibitor, Kun... 3435 0.0
22 BC004140|BC004140.1 Homo sapiens serine peptidase inhibitor, Kun... 3190 0.0
23 BT007425|BT007425.1 Homo sapiens serine protease inhibitor, Kuni... 3053 0.0
24 AY358969|AY358969.1 Homo sapiens clone DNA35880 HAI-1 (UNQ223) m... 2145 0.0
25 AY296715|AY296715.1 Homo sapiens hepatocyte growth factor activa... 1984 0.0
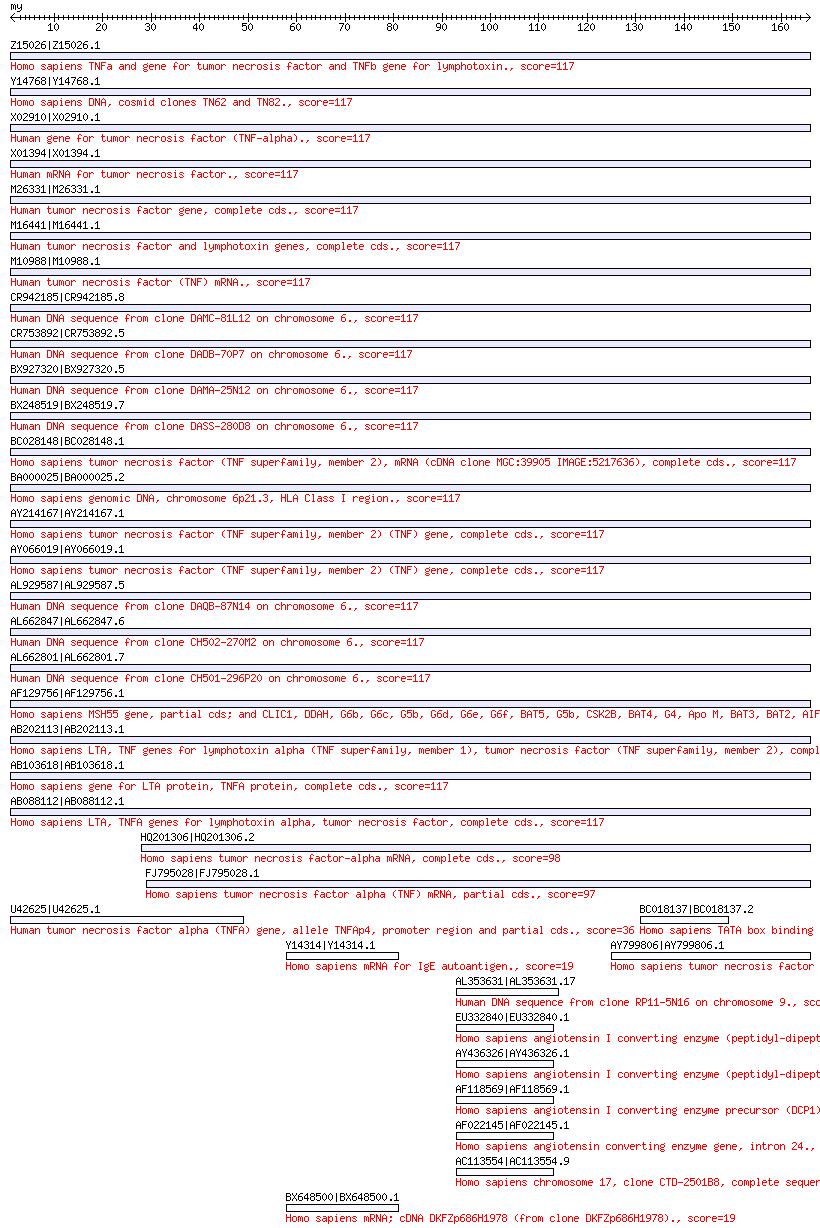
(後略)
BLAST search result image
BLAST search jobs submitted using WABI generate a graphical display of the search output.
Example:
WABI BLAST usage examples
Code example using Java
Example of code
- Example.java
- blast_condition.fasta
-
>my query sequence 1 CACCCTCTCTTCACTGGAAAGGACACCATGAGCACGGAAAGCATGATCCAGGACGTGGAA GCTGGCCGAGGAGGCGCTCCCCAGGAAGACAGCAGGGCCCCAGGGCTCCAGGCGGTGCTG GTTCCTCAGCCTCTTCTCCTTCCTGCTCGTGGCAGGCGCCGCCAC - blast_condition.txt
- Code
datasets ddbjall database hum program blastn parameters -v 100 -b 100 -e 10 -F F -W 11 format json result www - pom.xml
Execution example
- Preliminary preparation (Please run first)
-
$ wget 'http://sourceforge.jp/frs/redir.php?m=iij&f=%2Fjsonic%2F56583%2Fjsonic-1.3.0.zip' $ unzip jsonic-1.3.0.zip $ mv jsonic-1.3.0/jsonic-1.3.0.jar src/main/resources/ $ tree -F . ├── blast_condition.fasta ├── blast_condition.txt ├── pom.xml └── src/ └── main/ ├── java/ │ └── Example.java └── resources/ └── jsonic-1.3.0.jar - Build
-
$ mvn clean $ mvn compile $ mvn package $ mvn assembly:assembly -DdescriptorId=jar-with-dependencies - Execution procedure
-
$ java -classpath 'target/wabi-client-1.jar:target/wabi-client-1-jar-with-dependencies.jar:src/main/resources/jsonic-1.3.0.jar' Example
Code example using Perl
Example of code
- example.pl
- blast_condition.fasta
-
>my query sequence 1 CACCCTCTCTTCACTGGAAAGGACACCATGAGCACGGAAAGCATGATCCAGGACGTGGAA GCTGGCCGAGGAGGCGCTCCCCAGGAAGACAGCAGGGCCCCAGGGCTCCAGGCGGTGCTG GTTCCTCAGCCTCTTCTCCTTCCTGCTCGTGGCAGGCGCCGCCAC - blast_condition.txt
- Code
datasets ddbjall database hum program blastn parameters -v 100 -b 100 -e 10 -F F -W 11 format json result www
Execution example
- Preliminary preparation (Please run first)
-
$ cpan cpan[1] install JSON cpan[2]> install HTTP::Request::Common cpan[3]> install LWP::UserAgent cpan[4]> install HTTP::Status cpan[5]> quit $ tree -F . ├── blast_condition.fasta >├── blast_condition.txt └── example.pl - Execution procedure
-
$ perl example.pl
Code example using Ruby
Example of code
- example.rb
- blast_condition.fasta
-
>my query sequence 1 CACCCTCTCTTCACTGGAAAGGACACCATGAGCACGGAAAGCATGATCCAGGACGTGGAA GCTGGCCGAGGAGGCGCTCCCCAGGAAGACAGCAGGGCCCCAGGGCTCCAGGCGGTGCTG GTTCCTCAGCCTCTTCTCCTTCCTGCTCGTGGCAGGCGCCGCCAC - blast_condition.txt
- Code
datasets ddbjall
database hum
program blastn
parameters -v 100 -b 100 -e 10 -F F -W 11
format json
result www
Execution example
- Execution procedure
-
$ ruby example.rb - Actual example
Example of the execution result
- wabi_blast_2013-0606-1336-31-681-634313.txt
WABI BLAST Details
URI POST /blast (submit a search job)
This method appends a BLAST search to the job queue and
returns the Request ID.
The BLAST search criteria, method for notifying the result, and other
options are specified using HTTP parameters.
| Item | Description | |
|---|---|---|
| HTTP Method | POST | |
| URI | /blast | |
| HTTP Parameters | querySequence |
Search sequence data in multi-FASTA format Note: Increasing the number of sequences will not increase the degree of parallel processing. We recommend reducing the number of sequences searched for the web API, as this will better balance the load on the job management engine. |
datasets |
Dataset (example: "ddbjall") |
|
database |
database (example: "hum", "hum pri") |
|
program |
BLAST program (example: "blastn") |
|
parameters |
Other options (example: "-v 100 -b 100 -e 10 -F F -W 11") |
|
format |
response data format (example: "text", "json") |
|
result |
result retrieval method (example: "www", "mail") |
|
address |
Email address | |
| Request action | Add a new BLAST search job to the queue for processing. | |
| HTTP Response | If successful | Request ID |
| If failed | HTTP status 4xx | |
Server-side processing flow
- Check validity of input values.
If an invalid input value is found, then the processing is interrupted and HTTP status 400 Bad Request is returned. - Add the BLAST search into the job queue.
- Information about the job added to the queue is returned using the
specified format.
If the specified format is not available, then HTTP status 404 Not Found is returned.
Input validation check performed
|
datasets (optional) |
|
|---|---|
| database |
|
| program |
|
| format |
|
|
parameters (optional) |
|
| result |
|
| address |
|
Response data
- In the case of successful completion:
-
- A “successful” HTTP status code such as 200 will be returned.
- The field requestId will contain Request ID.
- Other values such as current-time and other request
information may be included in the response data; however,
formatting of these values may change in the future.
Please use the HTTP status code to determine the success or failure of the request.
- In the case of failed completion:
-
- A “client error” HTTP status code such as 400 will be returned.
- The key called error-messages included in the returned data will contain an error message indicating information such as the name of the parameter that caused input validation error.
- Other values including the cause of the error and other request
information may be included in the returned data; however,
formatting of these values may change in the future.
Please use the HTTP status code to determine the success or failure of the request.
Example request and response
Examples of input data
| HTTP Parameter | Input Value |
|---|---|
address |
"" |
database |
"hum" |
datasets |
"ddbjall" |
format |
"json" |
parameters |
" -v 100 -b 100 -e 10 -F F -W 11" |
program |
"blastn" |
querySequence |
|
result |
"www" |
Example response for a successfully completed request
HTTP Status 200
1 {
2 "requestId": "wabi_blast_1111-1111-1111-11-111-111111",
3 "program": "blastn",
4 "datasets": "ddbjall",
5 "database": "hum",
6 "parameters": " -v 100 -b 100 -e 10 -F F -W 11 ",
7 "current-time": "2013-01-01 12:34:56",
8 "start-time": "",
9 "current-state": ""
10 }
HTTP Status 200
1 <?xml version="1.0" ?>
2 <!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
3 <result>
4 <requestId>wabi_blast_1111-1111-1111-11-111-111111</requestId>
5 <program>blastn</program>
6 <datasets>ddbjall</datasets>
7 <database>hum</database>
8 <parameters> -v 100 -b 100 -e 10 -F F -W 11 </parameters>
9 <current-time>2013-01-01 12:34:56</current-time>
10 <start-time></start-time>
11 <current-state></current-state>
12 </result>
Example response data when there is an input validation error
HTTP Status 400
1 {
2 "status": "illegal-arguments",
3 "message": "Illegal arguments.",
4 "format": null,
5 "program": "blastn",
6 "datasets": "ddbjall",
7 "database": "hum",
8 "parameters": null,
9 "querySequence": ">my query sequence 1nCACCCTCTCTTCACTGGAAAGGACACCATGAGCACGGAAAGCATGATCCAGGACGTGGAAnGCTGGCCGAGGAGGCGCTCCCCAGGAAGACAGCAGGGCCCCAGGGCTCCAGGCGGTGCTGnGTTCCTCAGCCTCTTCTCCTTCCTGCTCGTGGCAGGCGCCGCCACn",
10 "result": "",
11 "address": null,
12 "current-time": "2013-01-01 12:34:56",
13 "error-messages": [
14 "Required: (result)"
15 ],
16 "error-message": "BAD_REQUEST (null)"
17 }
Usage Example
Example of the use of the REST client on Java Spring Framework:
URI GET /blast/{Request-ID}?info=status (query the status of a search job)
Returns the status of the job specified by Request ID.
| Item | Description | |
|---|---|---|
| HTTP Method | GET |
|
| URI |
"/blast/" + Request ID + HTTP Parameter
(example: "/blast/wabi_blast_1111-1111-1111-11-111-111111?info=status") |
|
| HTTP Parameters |
info (default value = "status") |
Job information type (example: "status", "result") |
format (default value = "text") |
Response data format (example: "text", "json") |
|
Server-Side Processing Flow
- Input values are validated on the server side.
If an input validation error is found, processing is interrupted, and the server returns HTTP status 400 Bad Request. - The server obtains job information corresponding to the specified
Request ID and returns the current
status.
HTTP status code 404 is returned if the search retention period has expired or if the job information cannot be found. - The server returns the obtained current job’s
status using the specified format.
However, the server returns HTTP status 404 Not Found if the specified format is invalid.
Input value validation check performed
| requestId |
|
|---|---|
| format |
|
| imageId |
|
| info |
|
Response Data
- In the case of successful completion:
-
- Returns a “successful” HTTP status code such as 200.
- The field status will contain information corresponding to “current status”.
- Other values such as current-time and other request
information may be included in the response data; however,
formatting of these values may change in the future.
Please use the HTTP status code to determine the success or failure of the request.
- In the case of failed completion:
-
- Returns a “client error” HTTP status code such as 400.
- The key called error-messages included in the returned data will contain an error message indicating such information as the name of the parameter that caused input validation error.
- Other values including cause of the error and other request
information may be included in the returned data; however,
formatting of these values may change in the future.
Please use the HTTP status code to determine the success or failure of the request.
Example request and response
Example input values
| HTTP Parameter | nput value |
|---|---|
| format | “json” |
| result | “www” |
| info | “status” |
Example response for a successfully completed request
HTTP Status 200
1 request-ID: wabi_blast_1111-1111-1111-11-111-111111
2 status: finished
3 current-time: 2013-01-01 12:34:56
4 system-info:
5 stdout
HTTP Status 200
1{
2 "request-ID": "wabi_blast_1111-1111-1111-11-111-111111",
3 "status": "finished",
4 "current-time": "2013-01-01 12:34:56",
5 "system-info": "nstdout"
6}
HTTP Status 200
1<?xml version="1.0" ?>
2<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
3<result>
4 <request-ID>wabi_blast_1111-1111-1111-11-111-111111</request-ID>
5 <status>finished</status>
6 <current-time>2013-01-01 12:34:56</current-time>
7 <system-info>stdout</system-info>
8</result>
Example response for a failed request
HTTP Status 400
1{
2 "status": "illegal-arguments",
3 "message": "Illegal arguments.",
4 "requestId": "wabi_blast_1111-1111-1111-11-111-111111",
5 "format": "json",
6 "imageId": null,
7 "info": "status",
8 "current-time": "2013-01-01 12:34:56",
9 "error-messages": [
10 "No such data: (requestId)"
11 ],
12 "error-message": "BAD_REQUEST (null)"
13}
Example Usage
Example using the REST client on Java Spring Framework:
URI GET /blast/{Request-ID}?info=request (obtain and confirm search criteria)
This will return the search criteria of a specified Request ID.
| Item | Description | |
|---|---|---|
| HTTP Method | GET |
|
| URI | “/blast/” + Request ID + “?info=request” + HTTP Parameters (example: “/blast/wabi_blast_1111-1111-1111-11-111-111111?info=request”) |
|
| HTTP Parameter | format(default value = “text”) |
Response data format (example: “text”, “json”) |
Server-Side Processing Flow
- Input values are validated on the server side.
If an input validation error is found, processing is interrupted, and the server returns HTTP status 400 Bad Request. - Information on the job specified by Request
ID is retrieved, and the search
criteria are returned.
HTTP status code 404 is returned if the search retention period has expired or if the job information cannot be found. - The server returns the current search criteria
in the specified format.
However, the server returns HTTP status code 404 Not Found if the specified format is invalid.
Input value validation check performed
| requestId |
|
|---|---|
| format |
|
| imageId |
|
| info |
|
Response Data
- In the case of successful completion:
-
- Returns a “successful” HTTP status code such as 200.
- Response data contain each parameter included when the BLAST search job was initially submitted.
- Please use the HTTP status code to determine the success or failure of the request.
- In case of failed completion:
-
- Returns a “client error” HTTP status code such as 400.
- The key called error-messages included in the returned data will contain an error message with information such as the name of the parameter that caused the input validation error.
- Please use the HTTP status code to determine the success or failure of the request.
Example request and response
Example input values:
| HTTP Parameters | Input values |
|---|---|
| format | “requestfile” |
| info | “request” |
Example response for a successfully completed request:
HTTP Status 200
1{
2 "address": "",
3 "database": "hum",
4 "datasets": "ddbjall",
5 "format": "text",
6 "parameters": " -v 100 -b 100 -e 10 -F F -W 11 ",
7 "program": "blastn",
8 "querySequence": ">my query sequence 1nCACCCTCTCTTCACTGGAAAGGACACCATGAGCACGGAAAGCATGATCCAGGACGTGGAAnGCTGGCCGAGGAGGCGCTCCCCAGGAAGACAGCAGGGCCCCAGGGCTCCAGGCGGTGCTGnGTTCCTCAGCCTCTTCTCCTTCCTGCTCGTGGCAGGCGCCGCCACn",
9 "result": "www"
10}
Example response for a failed request:
HTTP Status 404
1{
2 "Message": "Unexpected error ( Results of your request id have been NOT FOUND.)",
3 "requestId": "wabi_blast_1111-1111-1111-11-111-111111",
4 "format": "text",
5 "imageId": null,
6 "info": "result",
7 "current-time": "2013-01-01 12:34:56",
8 "error-messages": [
9 "No such data: (requestId)"
10 ],
11 "error-message": "NOT_FOUND (null)"
12}
Example Usage
Example using the REST client on Java Spring Framework:
URI GET /blast/{Request-ID}?info=result (retrieve search results)
Returns the search results for a specified Request ID.
| Item | Description | |
|---|---|---|
| HTTP Method | GET |
|
| URI |
"/blast/" + Request ID</code> + "?info=request" + HTTP Parameters
(example: "/blast/wabi_blast_1111-1111-1111-11-111-111111?info=request") |
|
| HTTP Parameter |
format (default value = "text") |
Response data format (example: "text", "json") |
Server-Side Processing Flow
- Input values are validated on the server side.
If an input validation error is found, processing is interrupted, and the server returns HTTP status code 400 Bad Request. - Information on the job specified by Request
ID is retrieved, and returns the search
result.
HTTP status code 400 or 404 is returned if the search retention period has expired or if the search processing has not yet completed. - The server returns the search result using the
specified format.
However, the server returns HTTP status code 404 Not Found if the specified format is invalid.
Input validation check performed
| requestId |
|
|---|---|
| format |
|
| imageId |
|
| info |
|
Response Data
- In the case of successful completion:
-
- Returns a “successful” HTTP status code such as 200.
- Returns the content of the output file generated through the BLAST search.
- Please use the HTTP status code to determine the success or failure of the request.
- In the case of failed completion:
-
- Returns a “client error” HTTP status code such as 400.
- The key called error-messages included in the returned data will contain an error message with information such as the name of the parameter that caused the input validation error.
- Please use the HTTP status code to determine the success or failures of the request.
Example request and response
Example of input data:
| HTTP Parameters | Input Value |
|---|---|
| format | “bigfile” |
| info | “result” |
Example response for a successfully completed request
HTTP Status 200
1 BLASTN 2.2.25 [Feb-01-2011]
2
3 Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
4 Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
5 "Gapped BLAST and PSI-BLAST: a new generation of protein database search
6 programs", Nucleic Acids Res. 25:3389-3402.
7
8 Query= AB000095|AB000095.1 Homo sapiens mRNA for hepatocyte growth
9 factor activator inhibitor, complete cds.
10 (1740 letters)
11
12 Database: hum
13 572,091 sequences; 5,019,832,159 total letters
14
15 Searching..................................................done
16
17 Score E
18 Sequences producing significant alignments: (bits) Value
19
20 AB000095|AB000095.1 Homo sapiens mRNA for hepatocyte growth fact... 3449 0.0
21 BC018702|BC018702.1 Homo sapiens serine peptidase inhibitor, Kun... 3435 0.0
22 BC004140|BC004140.1 Homo sapiens serine peptidase inhibitor, Kun... 3190 0.0
23 BT007425|BT007425.1 Homo sapiens serine protease inhibitor, Kuni... 3053 0.0
24 AY358969|AY358969.1 Homo sapiens clone DNA35880 HAI-1 (UNQ223) m... 2145 0.0
25 AY296715|AY296715.1 Homo sapiens hepatocyte growth factor activa... 1984 0.0
(後略)
Example response data when there is an input validation error
HTTP Status 400
1{
2 "Message": "Error ( Results of your request id have been NOT FOUND, or still running.)",
3 "requestId": "wabi_blast_1111-1111-1111-11-111-111111",
4 "format": "text",
5 "imageId": null,
6 "info": "result",
7 "current-time": "2013-01-01 12:34:56",
8 "error-messages": [
9 "No such data: (requestId)"
10 ],
11 "error-message": "NOT_FOUND (null)"
12}
Usage Example
Example of the use of the REST client on Java Spring Framework:
URI GET /blast/{Request-ID}?imageId={Image-ID} (retrieve image data generated from the search result)
This method returns the image data generated from the search result specified by RequestID.
| Item | Description | |
|---|---|---|
| HTTP Method | GET |
|
| URI |
"/blast/" + RequestID + "?imageId=" + Image ID + HTTP parameters
(example: "/blast/wabi_blast_1111-1111-1111-11-111-111111?imageId=1") |
|
| HTTP Parameters |
format (default value = "text") |
Response data format (example: "imagefile") |
Server-side processing flow
- Input values are validated on the server side.
If an invalid input value is found, then the processing is interrupted and HTTP status 400 Bad Request is returned. - Information about the job corresponding to the specified
RequestID is obtained, and the image
generated from the BLAST search is returned.
HTTP status code 400 or 404 is returned if the job information cannot be retrieved due to search retention period having expired or due to search job not having completed processing. - The server returns the image generated from the BLAST
search in the specified format.
However, the server returns HTTP status code 404 Not Found if the specified format is invalid.
Input validation check performed
| requestId |
|
|---|---|
| format |
|
| imageId |
|
| info |
|
Response Data
- In the case of successful completion:
-
- Returns a “successful” HTTP status code such as 200.
- Returns the image data generated from the BLAST search.
- Please use the HTTP status code to determine the success or failure of the request.
- In the case of failed completion:
-
- Returns a “client error” HTTP status code such as 400.
- The key called error-messages included in the returned data will contain an error message with information such as the name of the parameter that caused the input validation error.
- Please use the HTTP status code to determine the success or failure of the request.
Example request and response
Example input values
| HTTP Parameter | Input Value |
|---|---|
imageId |
"1" |
format |
"imagefile" |
info |
"" |
Example response for a successfully completed request
HTTP Status 200
(Image Data)
Example response for a failed request
HTTP Status 404
1 {
2 "Message": "Error ( Blast image file of your request id have been NOT FOUND, or still running.)",
3 "requestId": "wabi_blast_1111-1111-1111-11-111-111111",
4 "format": "text",
5 "imageId": null,
6 "info": "result",
7 "current-time": "2013-01-01 12:34:56",
8 "error-messages": [
9 "No such data: (requestId)"
10 ],
11 "error-message": "NOT_FOUND (null)"
12 }
Example Usage
Example using HttpURLConnection with Java:
URI GET /blast/help/{Help-Command} (View help information)
Returns the help information of WABI BLAST.
| Item | Description | |
|---|---|---|
| HTTP Method | GET |
|
| URI |
"/blast/help/" + Help-Command + HTTP Parameter
(example: "/blast/help/list_program?format=json") |
|
| HTTP Parameters |
format (default value = "text") |
Response data format (example: "text", "json") |
program |
BLAST program name (example: "blastn") |
|
Help-Command
| Help-Commands | Other HTTP Parameters | Help information that can be referenced |
|---|---|---|
list_datasets |
List of defined values for the datasets parameter | |
list_database |
List of defined values for the database parameter | |
list_program |
List of defined parameters for the BLAST program parameter | |
list_parameters |
program = BLAST program
|
List of defined optional parameters for the specified BLAST program |
list_format |
List of defined values for the response data parameter | |
list_result |
List of defined values for the result parameter | |
list_info |
パList of defined parameters for job information to be retrieved | |
Response data
- In the case of successful completion:
-
- Returns the requested help information
- In case of failed completion:
-
- Returns the list of valid Help-Commands and other information.
Example request and response
Example input values:
| Help-Command and other parameters | nput Values |
|---|---|
| Help-Command | “list_result” |
| format | “json” |
Example response for a successfully completed request:
1 {
2 "result": [
3 "www",
4 "mail"
5 ]
6 }
Example response for a failed request:
1 {
2 "help_commands": [
3 "list_datasets",
4 "list_database",
5 "list_program",
6 "list_parameters",
7 "list_format",
8 "list_result",
9 "list_info"
10 ],
11 "format": [
12 "text",
13 "json",
14 "xml"
15 ]
16 }
Example Usage
Example of the use of the REST client on Java Spring Framework:
WABI BLAST Parameters
requestId:Request ID
A string that is used to uniquely identify a BLAST search
job from those registered in WABI.
WABI returns Request ID as part of the response data
when the job is registered in the queue: this should be saved by the
program that uses WABI.
Request ID would be needed for performing the following tasks:
Example Request ID:
wabi_blast_1111-1111-1111-11-111-111111
Also refer to: BLAST Help Request ID
querySequence: Query sequence data
- Query sequence must be in FASTA format.
- In order to assign a name to a sequence, include a line starting with “>” before each sequence.
- If there are multiple sequences to be queried, then sequence names
are mandatory in order to distinguish between the sequences
(multi-FASTA format).
Note: Increasing the number of sequences will not increase the degree of parallel processing. We recommend reducing the number of sequences searched using the web API, considering the load balancing that is applied by the job management engine. - Sequence name is not required if you only have one sequence to search.
Example sequence in FASTA format
>my query sequence 1
CACCCTCTCTTCACTGGAAAGGACACCATGAGCACGGAAAGCATGATCCAGGACGTGGAA
GCTGGCCGAGGAGGCGCTCCCCAGGAAGACAGCAGGGCCCCAGGGCTCCAGGCGGTGCTG
GTTCCTCAGCCTCTTCTCCTTCCTGCTCGTGGCAGGCGCCGCCAC
Example of multiple sequences (multi-FASTA format)
>my query sequence 1
CACCCTCTCTTCACTGGAAAGGACACCATGAGCACGGAAAGCATGATCCAGGACGTGGAA
GCTGGCCGAGGAGGCGCTCCCCAGGAAGACAGCAGGGCCCCAGGGCTCCAGGCGGTGCTG
GTTCCTCAGCCTCTTCTCCTTCCTGCTCGTGGCAGGCGCCGCCAC
>my query sequence 2
GGCCAGGGCACCCAGTCTGAGAACAGCTGCACCCGCTTCCCAGGCAACCTGCCTCACATG
CTTCGAGACCTCCGAGATGCCTTCAGCAGAGTGAAGACTTTCTTTCAAATGAAGGATCAG
CTGGACAACATATTGTTAAAGGAGTCCTTGCTGGAGGACTTTAAG
>my query sequence 3
ATGGGTCTCACCTCCCAACTGCTTCCCCCTCTGTTCTTCCTGCTAGCATGTGCCGGCAAC
TTTGCCCACGGACACAACTGCCATATCGCCTTACGGGAGATCATCGAAACTCTGAACAGC
CTCACAGAGCAGAAGACTCTGTGCACCAAGTTGACCATAACGGAC
Valid search results may not be obtained with very long sequences or if
there are too many sequences for the following reasons.
In such cases, please try reducing the number of query sequences or
making sequences shorter.
- Search may end abnormally due to memory running out.
- Search may time out due to the search taking too long.
Also refer to: BLAST Help Query name, Query sequence
datasets:Datasets
Datasets are available to assist in completing the query form on the web interface, but they are not currently used in WABI.
Valid input values that can be specified for datasets and their
respective meanings are explained below.
Note: Please use the help API GET
/blast/help/{Help-Command} to reference the most
up-to-date information.
| Dataset value | Explanation |
|---|---|
ddbjall |
DDBJ ALL (DDBJ periodical release + daily updates) |
ddbjnew |
DDBJ New (DDBJ daily updates) |
16S_rRNA |
16S rRNA (Prokaryotes) |
uniprot_all |
UniProt (Swiss-Prot + TrEMBL) |
uniprot_sprot |
UniProt (Swiss-Prot) |
uniprot_trembl |
UniProt (TrEMBL) |
patent_protein |
Patent |
dadall |
DAD (periodical release + daily updates) |
dadnew |
DAD (daily updates) |
refseq_na |
RefSeq NA |
refseq_aa |
RefSeq AA |
Also refer to: BLAST HELP Nucleotide (DATABASE, DIVISION)
database:Database
Nucleotide Sequence Database
Example nucleotide sequence database values and their corresponding
explanations are listed in the table below.
Note: Please use the help API GET
/blast/help/{Help-Command} to reference the most
recently updated information.
| Explanation | Database value | |
|---|---|---|
| DDBJ ALL | DDBJ periodical release + daily updates | (Refer to table below) |
| DDBJ New | DDBJ daily updates | (Add prefix “new_” to the values below) |
| 16S rRNA | 16S rRNA from DDBJ periodical release | 16S_rRNA |
| RefSeq NA | RefSeq (Genomics + RNA) | (Refer to table below) |
DDBJ ALL, DDBJ NEW Database value
| Standard divisions | ||
|---|---|---|
hum, new_hum |
Human | human |
pri, new_pri |
Primates | primates other than human |
rod, new_rod |
Rodents | rodents |
mam, new_mam |
Mammals | mammals other than human, primates and rodents |
vrt, new_vrt |
Vertebrates | vertebrates other than human, primates, rodents and mammals |
inv, new_inv |
Invertebrates | invertebrates |
pln, new_pln |
Plants | plants |
bct, new_bct |
Bacteria | bacteria |
vrl, new_vrl |
Viruses | viruses |
phg, new_phg |
Phages | phages |
syn, new_syn |
Synthetic DNAs | synthetic DNAs (SYN) |
env, new_env |
ENV | environmental samples(environmental samples) |
| High throughput divisions | ||
|---|---|---|
htc, new_htc |
HTC | High Throughput cDNAs |
htg, new_htg |
HTG | High Throughput Genomic sequences |
tsa, new_tsa |
TSA | Transcriptome Shotgun Assembly |
| EST divisions | ||
|---|---|---|
est_atha, new_est_atha |
A.thaliana | Arabidopsis thaliana (thale cress) |
est_btra, new_est_btra |
B.taurus | Bos taurus (cattle) |
est_cele, new_est_cele |
C.elegans | Caenorhabditis elegans (nematode worm) |
est_crei, new_est_crei |
C.reinhardtii | Chlamydomonas reinhardtii (Chlamydomonas:green algae) |
est_cint, new_est_cint |
C.intestinalis | Ciona intestinalis (vase tunicate) |
est_drer, new_est_drer |
D.rerio | Danio rerio (zebrafish) |
est_ddis, new_est_ddis |
D.discoideum | Dictyostelium discoideum (soil-living amoeba) |
est_dmel, new_est_dmel |
D.melanogaster | D.melanogaster (fruit fly) |
est_ggal, new_est_ggal |
G.gallus | Gallus gallus (chicken) |
est_gmax, new_est_gmax |
G.max | Glycine max (soybean) |
est_hum, new_est_hum |
H.sapiens | Homo sapiens (human) |
est_hvul, new_est_hvul |
H.vulgare | Hordeum vulgare (Barley) (incl. subspecies) |
est_mtru, new_est_mtru |
M.truncatula | Medicago truncatula (Barrel Medic) (incl. mixed library) |
est_mous, new_est_mous |
M.musculus | Mus musculus (Mouse) |
est_osat, new_est_osat |
O.sativa | Oryza sativa (incl. subspecies rank) |
est_rnor, new_est_rnor |
R.norvegicus | Rattus norvegicus (Rat) (incl. Rattus sp.) |
est_slyc, new_est_slyc |
S.lycopersicum | Solanum lycopersicum (tomato) |
est_taes, new_est_taes |
T.aestivum | Triticum aestivum (bread wheat) |
est_xlae, new_est_xlae |
X.laevis | Xenopus laevis (african clawed frog) |
est_xtro, new_est_xtro |
X.tropicalis | Xenopus tropicalis (western clawed frog) |
est_zmay, new_est_zmay |
Z.mays | Zea mays (maize) |
est_rest, new_est_rest |
Others | Others |
| Others | ||
|---|---|---|
pat, new_pat |
Patent | patent (PAT) |
una, new_una |
Unannotated Seq | unannotated sequences (UNA) |
gss, new_gss |
GSS | genome survey sequences |
sts, new_sts |
STS | sequence tagged sites |
Refseq NA Database value
| RefSeq NA | |
|---|---|
refseq-genomic-fungi, refseq-rna-fungi |
Fungi |
refseq-genomic-invertebrate, refseq-rna-invertebrate |
Invertebrate |
refseq-genomic-microbial, refseq-rna-microbial |
Microbial |
refseq-genomic-mitochondrion, refseq-rna-mitochondrion |
Mitochondrion |
refseq-genomic-plant, refseq-rna-plant |
Plant |
refseq-genomic-plasmid, refseq-rna-plasmid |
Plasmid |
refseq-genomic-plastid, refseq-rna-plastid |
Plastid |
refseq-genomic-protozoa, refseq-rna-protozoa |
Protozoa |
refseq-genomic-vertebrate_mammalian, refseq-rna-vertebrate_mammalian |
Vertebrate Mammalian |
refseq-genomic-vertebrate_other, refseq-rna-vertebrate_other |
Vertebrate Other |
refseq-genomic-viral, refseq-rna-viral |
Viral |
refseq-genomic |
RefSeq Genomic (ALL) Periodical Release |
refseq-rna |
RefSeq RNA (ALL) Periodical Release |
refseq-na-daily |
RefSeq Daily Updates |
refseq-na-all |
RefSeq ALL (Periodical Release + Daily Updates) |
refseq-model-rna-B_taurus |
B. taurus |
refseq-model-rna-D_rerio |
D. rerio |
refseq-model-rna-H_sapiens, refseq-model-genomic-H_sapiens |
H. sapiens |
refseq-model-rna-M_musculus |
M. musculus |
refseq-model-rna-R_norvegicus |
R. norvegicus |
refseq-model-rna-X_tropicalis |
X. tropicalis |
Amino Acid Sequence Databases
Example amino acid sequence database values and their corresponding explanations are listed in the table below.
| Explanation | Database value | |
|---|---|---|
| UniProt (Swiss-Prot + TrEMBL) | Swiss-Prot + TrEMBL | uniprot_all |
| UniProt (Swiss-Prot) | Swiss-Prot | uniprot_sprot |
| UniProt (TrEMBL) | TrEMBL | uniprot_trembl |
| Patent | JPO,EPO,USPTO,KIPO | jpop, epop, usptop, kipop |
| DAD ALL | periodical release + daily updates | (Refer to table below) |
| DAD NEW | daily updates | (Refer to table below) |
| RefSeq AA | RefSeq (Protein) | (Refer to table below) |
DAD ALL, DAD NEW Database value
| Standard divisions | ||
|---|---|---|
dad_hum, dad_new_hum |
Human | human |
dad_pri, dad_new_pri |
Primates | primates other than human |
dad_rod, dad_new_rod |
Rodents | rodents |
dad_mam, dad_new_mam |
Mammals | mammals other than human,primates and rodents |
dad_vrt, dad_new_vrt |
Vertebrates | vertebrates other than human,primates, rodents and mammals |
dad_inv, dad_new_inv |
Invertebrates | invertebrates |
dad_pln, dad_new_pln |
Plants | plants |
dad_bct, dad_new_bct |
Bacteria | bacteria |
dad_vrl, dad_new_vrl |
Viruses | viruses |
dad_phg, dad_new_phg |
Phages | phages |
dad_syn, dad_new_syn |
Synthetic DNAs | synthetic DNAs (SYN) |
dad_env, dad_new_env |
General | environmental samples |
| High throughput divisions | ||
|---|---|---|
dad_htc, dad_new_htc |
HTC | High Throughput cDNAs |
dad_htg, dad_new_htg |
HTG | High Throughput Genomic sequences |
dad_tsa, dad_new_tsa |
TSA | Transcriptome Shotgun Assembly |
| EST divisions | ||
|---|---|---|
dad_est_atha, dad_new_est_atha |
A.thaliana | Arabidopsis thaliana (thale cress) |
dad_est_btra, dad_new_est_btra |
B.taurus | Bos taurus (cattle) |
dad_est_cele, dad_new_est_cele |
C.elegans | Caenorhabditis elegans (nematode worm) |
dad_est_crei, dad_new_est_crei |
C.reinhardtii | Chlamydomonas reinhardtii (Chlamydomonas:green algae) |
dad_est_cint, dad_new_est_cint |
C.intestinalis | Ciona intestinalis (vase tunicate) |
dad_est_drer, dad_new_est_drer |
D.rerio | Danio rerio (zebrafish) |
dad_est_ddis, dad_new_est_ddis |
D.discoideum | Dictyostelium discoideum (soil-living amoeba) |
dad_est_dmel, dad_new_est_dmel |
D.melanogaster | D.melanogaster (fruit fly) |
dad_est_ggal, dad_new_est_ggal |
G.gallus | Gallus gallus (chicken) |
dad_est_gmax, dad_new_est_gmax |
G.max | Glycine max (soybean) |
dad_est_hum, dad_new_est_hum |
H.sapiens | Homo sapiens (human) |
dad_est_hvul, dad_new_est_hvul |
H.vulgare | Hordeum vulgare (Barley) (incl. subspecies) |
dad_est_mtru, dad_new_est_mtru |
M.truncatula | Medicago truncatula (Barrel Medic) (incl. mixed library) |
dad_est_mous, dad_new_est_mous |
M.musculus | Mus musculus (Mouse) |
dad_est_osat, dad_new_est_osat |
O.sativa | Oryza sativa (incl. subspecies rank) |
dad_est_rnor, dad_new_est_rnor |
R.norvegicus | Rattus norvegicus (Rat) (incl. Rattus sp.) |
dad_est_slyc, dad_new_est_slyc |
S.lycopersicum | Solanum lycopersicum (tomato) |
dad_est_taes, dad_new_est_taes |
T.aestivum | Triticum aestivum (bread wheat) |
dad_est_xlae, dad_new_est_xlae |
X.laevis | Xenopus laevis (western clawed frog) |
dad_est_xtro, dad_new_est_xtro |
X.tropicalis | Xenopus tropicalis (african clawed frog) |
dad_est_zmay, dad_new_est_zmay |
Z.mays | Zea mays (maize) |
dad_est_rest, dad_new_est_rest |
Others | Others |
| Others | ||
|---|---|---|
dad_pat, dad_new_pat |
Patent | patent (PAT) |
dad_una, dad_new_una |
Unannotated Seq | unannotated sequences (UNA) |
dad_gss, dad_new_gss |
GSS | genome survey sequences |
dad_sts, dad_new_sts |
STS | sequence tagged sites |
Refseq AA Database value
| RefSeq AA | ||
|---|---|---|
refseq-protein-fungi |
Fungi | |
refseq-protein-invertebrate |
Invertebrate | |
refseq-protein-microbial |
Microbial | |
refseq-protein-mitochondrion |
Mitochondrion | |
refseq-protein-plant |
Plant | |
refseq-protein-plasmid |
Plasmid | |
refseq-protein-plastid |
Plastid | |
refseq-protein-protozoa |
Protozoa | |
refseq-protein-vertebrate_mammalian |
Vertebrate Mammalian | |
refseq-protein-vertebrate_other |
Vertebrate Other | |
refseq-protein-viral |
Viral | |
refseq-protein |
RefSeq Protein (ALL) Periodical Release | |
refseq-aa-daily |
RefSeq Protein Daily Updates | |
refseq-aa-all |
RefSeq Protein ALL (Periodical Release + Daily Updates) | |
refseq-model-protein-B_taurus |
B. taurus | |
refseq-model-protein-D_rerio |
D. rerio | |
refseq-model-protein-H_sapiens |
H. sapiens | |
refseq-model-protein-M_musculus |
M. musculus | |
refseq-model-protein-R_norvegicus |
R. norvegicus | |
refseq-model-protein-X_tropicalis |
X. tropicalis |
Also refer to: BLAST HELP Nucleotide (DATABASE, DIVISION)
program:BLAST Program
You can choose from the following BLAST programs depending on the
analysis being performed.
Note: Please use the help API GET
/blast/help/{Help-Command} to reference the most
recently updated information.
| BLAST Program | query | Data Base | Explanation |
|---|---|---|---|
| megablast | nucleotide | nucleotide | Aligning your nucleotide sequence with nucleotide sequence database. When you want to perform a homology search with long length of nucleotide sequence, results are provided faster than blastn program. |
| blastn | nucleotide | nucleotide | Aligning your nucleotide sequence with nucleotide sequence database. |
| tblastn | amino acid | nucleotide | Aligning your amino acid sequence with nucleotide sequence database by translating database sequences taking into account all six possible open reading frames. |
| tblastx | nucleotide | nucleotide | Aligning your nucleotide sequence with nucleotide sequence database by translating both sequences taking into account all six possible open reading frames. |
| blastp | amino acid | amino acid | Aligning your amino acid sequence with amino acid seque nce database. |
| blastx | nucleotide | amino acid | Aligning your nucleotide sequence with amino acid sequence database by translating your sequence taking into account all six possible open reading frames. |
Also refer to: BLAST HELP Program
parameters:BLAST program options
BLAST program options that can be specified are as follows:
Note: Please use the help API GET
/blast/help/{Help-Command} to reference the most
recently updated information.
Combinations of these options can be specified with corresponding values separated by spaces.
| Options | BLAST Program | Explanation |
|---|---|---|
-A N |
All programs | Multiple Hits window size; generally defaults to 0 (for single-hit extensions), but defaults to 40 when using discontiguous templates. |
-B N |
All programs except "megablast" |
Number of concatenated queries, in blastn or tblastn mode |
-C X |
All programs except "megablast" |
Use composition-based statistics for blastp or tblastn: T, t, D, or d Default (equivalent to 1 for blast2 and blastall_old and to 2 for blastall and blastcl3) 0, F, or f No composition-based statistics 1 Composition-based statistics as in NAR 29:2994-3005, 2001 2 Composition-based score adjustment as in Bioinformatics 21:902-911, 2005, conditioned on sequence properties 3 Composition-based score adjustment as in Bioinformatics 21:902-911, 2005, unconditionally When enabling statistics in blastall, blastall_old, or blastcl3 (i.e., not blast2), appending u (case-insensitive) to the mode enables use of unified p-values combining alignment and compositional p-values in round 1 only. |
-D N |
All programs except "megablast" |
Translate sequences in the database according to genetic code N in /usr/share/ncbi/data/gc.prt (default is 1; only applies to tblast*) |
"megablast" |
Type of output: 0 alignment endpoints and score 1 all ungapped segments endpoints 2 traditional BLAST output (default) 3 tab-delimited one line format 4 incremental text ASN.1 5 incremental binary ASN.1 |
|
-E N |
"megablast" |
Extending a gap costs N (-1 invokes default behavior) |
All programs except "megablast" |
Extending a gap costs N (-1 invokes default behavior: non-affine if greedy, 2 otherwise) | |
-Fstr |
All programs | Filter options for DUST or SEG; defaults to T for bl2seq, blast2, blastall, blastall_old, blastcl3, and megablast, and to F for blastpgp, impala, and rpsblast. |
-G N |
"megablast" |
Opening a gap costs N (-1 invokes default behavior) |
All programs except "megablast" |
Opening a gap costs N (-1 invokes default behavior: non-affine if greedy, 5 if using dynamic programming) | |
-HN |
"megablast" |
Maximal number of HSPs to save per database sequence (default is 0, unlimited) |
-I |
All programs | Show GIs in deflines |
-J |
All programs | Believe the query defline |
-KN |
All programs except "megablast" |
Number of best hits from a region to keep. Off by default. If used a value of 100 is recommended. Very high values of -v or -b are also suggested. |
-Lstart, stop |
All programs | Location on query sequence (for rpsblast, only valid in blastp mode) |
-Mstr |
All programs except "megablast" |
Use matrix str (default = BLOSUM62) |
-MN |
"megablast" |
Maximal total length of queries for a single search (default = 5000000) |
-NN |
"megablast" |
Type of a discontiguous word template: 0 coding (default) 1 optimal 2 two simultaneous |
-P N |
All programs except "megablast" |
Set to 1 for single-hit mode or 0 for multiple-hit mode (default). Does not apply to blastn. |
"megablast" |
Maximal number of positions for a hash value (set to 0 [default] to ignore) | |
-QN |
All programs except "megablast" |
Translate query according to genetic code N in /usr/share/ncbi/data/gc.prt (default is 1) |
-R |
"megablast" |
Report the log information at the end of output |
-SN |
All programs | Query strands to search against database for blastn, blastx, tblastx: 1 top 2 bottom 3 both (default) |
-T |
All programs | Produce HTML output |
-U |
All programs | Use lower case filtering for the query sequence |
-V |
All programs | Force use of legacy engine |
-WN |
All programs | Use words of size N (length of best perfect match; zero invokes default behavior, except with megablast, which defaults to 28, and blastpgp, which defaults to 3. The default values for the other commands vary with "program": 11 for blastn, 28 for megablast, and 3 for everything else.) |
-XN |
All programs | X dropoff value for gapped alignment (in bits) (zero invokes default behavior, except with megablast, which defaults to 20, and rpsblast and seedtop, which default to 15. The default values for the other commands vary with "program": 30 for blastn, 20 for megablast, 0 for tblastx, and 15 for everything else.) |
-YX |
All programs | Effective length of the search space (use zero for the real size) |
-ZN |
All programs | X dropoff value for final [dynamic programming?] gapped alignment in bits (default is 100 for blastn and megablast, 0 for tblastx, 25 for others) |
-bN |
All programs | Number of database sequences to show alignments for (B) (default is 250) |
-eX |
Expectation value (E) (default = 10.0) | |
-fX |
All programs except "megablast" |
Threshold for extending hits, default if zero: 0 for blastn and megablast, 11 for blastp, 12 for blastx, and 13 for tblasn and tblastx. |
-f |
"megablast" |
Show full IDs in the output (default: only GIs or accessions) |
-g F |
All programs except "megablast" |
Do not perform gapped alignment (N/A for tblastx) |
"megablast" |
Make discontiguous megablast generate words for every base of the database (mandatory with the current BLAST engine) |
|
-lstr |
All programs | Restrict search of database to list of GI's [String] |
-mN |
All programs | alignment view options: 0 pairwise (default) 1 query-anchored showing identities 2 query-anchored, no identities 3 flat query-anchored, show identities 4 flat query-anchored, no identities 5 query-anchored, no identities and blunt ends 6 flat query-anchored, no identities and blunt ends 7 XML Blast output (not available for impala) 8 tabular (not available for impala) 9 tabular with comment lines (not available for impala) 10 ASN.1 text (not available for impala or rpsblast) 11 ASN.1 binary (not available for impala or rpsblast) |
-n |
All programs except "megablast" |
megablast search |
"megablast" |
Use non-greedy (dynamic programming) extension for affine gap scores | |
-pX |
"megablast" |
Identity percentage cut-off (default = 0) |
-qN |
All programs | Penalty for a nucleotide mismatch (blastn only) (default = -10 for seedtop, -3 for everything else) |
-rN |
All programs | Reward for a nucleotide match (blastn only) (default = 10 for seedtop, -10 for everything else) |
-s |
All programs except "megablast" |
Compute locally optimal Smith-Waterman alignments. For blastall, blastall_old, and blastcl3, this is only available in gapped tblastn mode. |
-sN |
"megablast" |
Minimal hit score to report (0 for default behavior) |
-t N |
All programs except "megablast" |
Length of a discontiguous word template (the largest intron allowed in a translated nucleotide sequence when linking multiple distinct assignments; default = 0; negative values disable linking for blastall, blastall_old, and blastcl3.) |
"megablast" |
Length of a discontiguous word template (contiguous word if 0 [default]) | |
-vN |
All programs | Number of one-line descriptions to show (V) (default = 500) |
-wN |
All programs except "megablast" |
Frame shift penalty (OOF algorithm for blastx) |
-yX |
All programs except "megablast" |
X dropoff for ungapped extensions in bits (0.0 invokes default behavior: 20 for blastn, 10 for megablast, and 7 for all others.) |
-yN |
"megablast" |
X dropoff value for ungapped extension (default is 10) |
-zN |
All programs | Effective length of the database (use zero for the real size) |
Example BLAST program option:
-v 100 -b 100 -e 10 -F F -W 11
Also refer to: BLAST HELP Optional Parameters
format:Response data format
You can specify the following options to select the WABI response data
format.
Note: Please use the help API GET /blast/help/{Help-Command} to
reference the most recently updated information.
| Response data format | Explanation | Media Type |
|---|---|---|
text |
Plain text | text/plain; charset=utf-8 |
json |
JSON format | application/json; charset=utf-8 |
xml |
XML text | text/xml; charset=utf-8 |
bigfile |
Used when retrieving data that is output to a file, such as search results as plain text. | text/plain; charset=utf-8 |
imagefile |
Image data | image/png |
requestfile |
Used when retrieving data that is output to a file, such as search results as JSON text. | application/json; charset=utf-8 |
Note: If WABI cannot generate response data in the specified format, then it is considered an invalid input value and returns an HTTP error code.
result:Result retrieval method
The method for retrieving results can be specified from one of the
following:
Note: Please use the help API GET
/blast/help/{Help-Command} to reference the most
recently updated information.
| Retrieval Method | Explanation |
|---|---|
www |
Submit a request to a URL for retrieving a result, and receive the result as the response data to the request. |
mail |
Result is sent to the specified email address. |
address:Email address
The email address to which the results will be sent.
info:The type of job information being referenced
The following types of information can be retrieved for a submitted
search job.
Note: Please use the help API GET
/blast/help/{Help-Command} to reference the most
up-to-date information.
| Information Type | Explanation |
|---|---|
status |
Job status |
result |
Search results |
request |
Search criteria specified when the job was submitted |
imageId:The ID of the image associated with a search output
This ID is used to retrieve the image data generated as part of a search.
It is required for the following case:
Example ID for an image generated by search output:
1
Other Information
Search results are retained for 7 days.
(Refer to “Search result retention period” 「Request ID and BLAST
result」)
You can obtain the processed search result at any
time during this period by submitting a GET request specifying the
Request ID.