Dr. Sugawara, has been committing to the worldwide activities of the WFCC over many years in particular to Newsletter editing, training, information dissemination, providing statistics on collections worldwide and not least the direction of the World Data Centre for Microorganisms(WDCM). He constructs and maintains the WDCM web site on his own laboratory web site. Through this web site, he continued to provide the scientific community for the microorganism researchers, as well as spread the useful information to all over the world. More recently he also contributed to establishment of OECD Global Biodiversity Information Facility (GBIF)) and the design of OECD Global Biological Resource Centre Network.
Appreciating his contribution to the WFCC for these years, the WFCC decided to elect him to the Honorary Membership of the WFCC and present the WFCC Medal, on the occasion of his retirement from the National Institute of Genetics(NIG). For the presentation ceremony, Dr. Ken-Ichiro Suzuki, Vice-President of the WFCC (and director of National Institute of Technology and Evaluation(NITE)) visited Mishima-city (where NIG is located) on March 24, 2008. Dr. Suzuki read for the message of Dr. David Smith, President of the WFCC, and handed the award medal to Dr. Sugawara. Many DDBJ staffs congratulated his honor at the ceremony, together with his family.
 |
 |
 |
| Dr. Suzuki presented the certificate | certificate | WFCC medal (small red box) |
- DDBJ Rel.72
- File name: ddbjcon1.seq.gz, ddbjcon1.insd_xml.gz
- Accession number: reference(3,514 data)
- Current status: Correction of DDBJ rel.73 (release on March 27,2008)
- DAD Rel.42
- File name: ddbjcon1.DAD.gz
- Accession number: reference(30,434 data)
- Current status: Correction of DAD rel.43 (release in the first of Aplil,2008)
which had been submitted by RIKEN Genome Sciences Center.
The correct versions were "BA000010.8" and "BA000044.2" respectively, but they appeared as "BA000010.9" and "BA000044.3".
- Relevant accession numbers: BA000010, BA000044
- Affected DDBJ services and period:
- - getentry /ARSA: 2008/02/19 - 2008/03/05
- - Anonymous FTP site: ftp://ftp.ddbj.nig.ac.jp/ddbj_database/ddbjnew/contig/
- DDBJNEWr72.086.CON.gz: Including the enties with invalid sequence version number ( BA000010.9, BA000044.3 ).
- DDBJNEWr73.014.CON.gz: Including the fixed enties.
- - getentry /ARSA: 2008/02/19 - 2008/03/05
- Cause: A software problem in the flatfile making process.
- Current status: We have already fixed and deleted the invalid version entries
in 2008/03/08 on the getentry and annonymous FTP (Daily update).
- Changed directory names and structure
- There were divided into ddbj_database (the DDBJ data) and mirror_database (the other databases data) directories though the DDBJ data and the other databases data were put together in the database under a top directory so far.
---PAST---
ftp://ftp.ddbj.nig.ac.jp/database/ :from the DDBJ data and from the other databases
---NOW---
ftp://ftp.ddbj.nig.ac.jp/ddbj_database/ :from the DDBJ data
ftp://ftp.ddbj.nig.ac.jp/mirror_database/ :from the other databases data - Kind README
- Detailed data in the ddbj_database¡¤we made README.TXT
- Notice -
The Old directory(database) will be maintained one month. When regularaly watch, please change to the new directories.
Pict1. A top position of anonymous FTP(2008.2.26)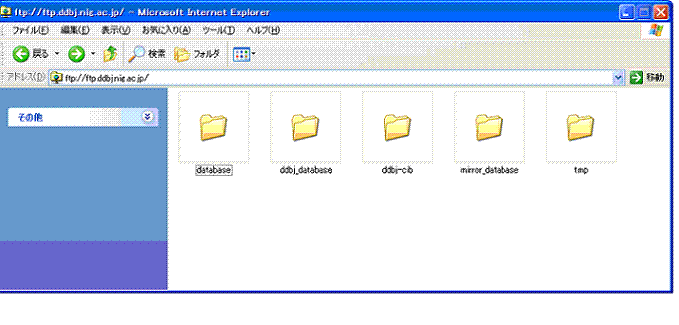
Pict2. A part of README.TXT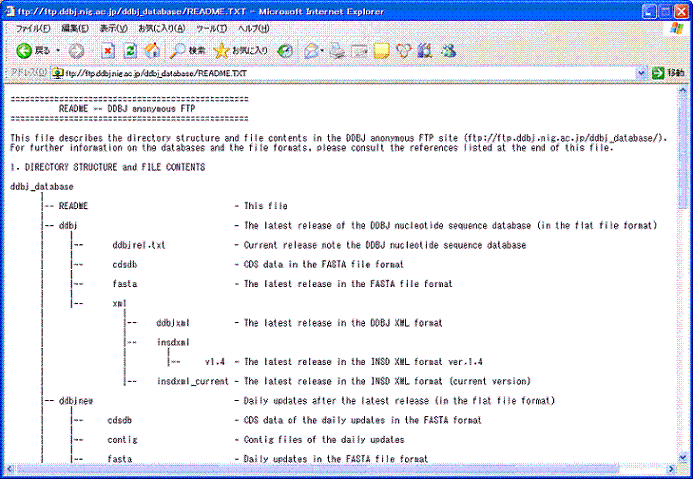
Number of nucleotide sequence: 168,562 entries (93,982,299 base-pair)
Number of amino acid sequence: 113,555 entries (16,499,605 amino-acid)
---Note---
Besides the data of KIPO, International Nucleotide Sequence Database (INSD) (collaboration of DDBJ, EMBL and GenBank) distribute other patent data that was accepted by Japan Patent Office (JPO), European Patent Office (EPO) and United States Patent and Trademark Office (USPTO).
For details, please refer to "Nucleotide Sequences Included Patent Applications" section of Patent, Intellectual Property and Priority page.
The prefix of the accession number of KIPO patent is "DI". Please refer from the following to the example of the open to the public data.
(ex) DI000001
---Note---
So, the prefixes for patent related data are;
JPO: E, BD, DD, DJ
EPO: A, AX, CQ, CS, FB
USPTO: I, AR, DZ, EA
KIPO: DI
The following figure shows summary of TOP 10 of number of ORGANISM in KIPO entries.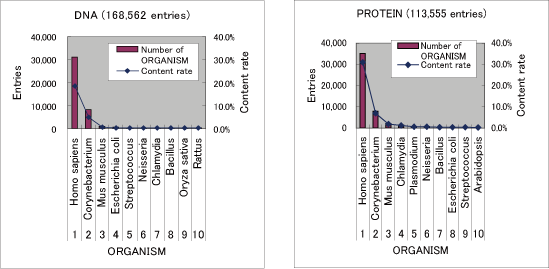
This announce introduced the applied data from KIPO that distributed first time. In the future, DDBJ and KIPO will build a mechanism that regular acquisition and distribution KIPO data.
この度公開された,韓国特許庁 (KIPO) からの特許出願塩基配列データは以下の通りです。
| entry number | accession number | |
| DNA | 168,562 | DI000001-DI168562 |
| PROTEIN | 113,555 | DI500001-DI613555 |
| TOTAL | 282,117 |
どちらも getentry より検索可能です。
DDBJ, EMBL, and GenBank take the publishable patent DNA data into the INSD,
mediating JPO, EPO and USPTO, respectively. And also from this February 2008,DDBJ started to take into and release patent related DNA data by KoreanIntellectual Property Office (KIPO)
Following DNA and protein data from KIPO was released by DDBJ;
| entry number | accession number | |
| DNA | 168,562 | DI000001-DI168562 |
| PROTEIN | 113,555 | DI500001-DI613555 |
| TOTAL | 282,117 |
These entries are searchable by getentry.
Note: DDBJ has been functioning as one of the International Nucleotide Sequence
Database Collaboration, including EBI (European Bioinformatics Institute; responsible
for the EMBL database) in Europe and NCBI (National Center for Biotechnology).
Information; responsible for GenBank database) in the USA as the two other members.
The old directory "database" was divided to "ddbj_database" (DDBJ origin data)
and "mirror_database" (mirror data of another database).
For details of "ddbj_database", please see README.TXT in this directory.
Old directory (database) is maintained for one month, If you watch
regularlly, please change to new directory.
The service was resumed at 16:22 on Feb. 26. Thank you for you cooperation.
You might recognize that you can hold your entries in a certain period or until the relevant paper are published. DDBJ should not disclose these Hold-Until-Published entries to the public. To retain them inside DDBJ, DDBJ stores these entries in a separate server from servers for data dissemination, e.g. getentry server.
In the daily data release procedure shown in the following figure, DDBJ convert entries into the flat file format from the server for in-house data management to transfer them to other servers for such services to the public as getentry, ARSA, homology search and anonymous FTP
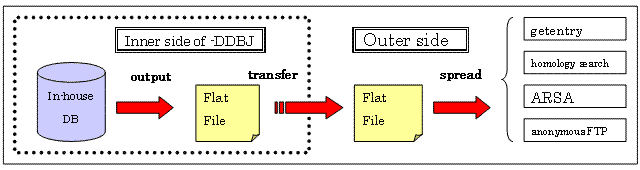
This month (January 2008), DDBJ released about 1,500,000 entries only in two days; about 1,000,000 entries in the first day, and about 500,000 entries in the second day. It is to be noted that this process was completed as smoothly as designed. With the previous system, we were able to process only 350,000 entries in a day, namely, we had to spend more than 4 days to complete 1.5M entries.
Although the mission was completed this time, we learned some lessons to improve the procedure further. You may claim that "2 days" is not quick enough. The system engineers in charge started the discussion to improve the concordance of several servers to gain higher throughput than ever.
We would appreciate it very much, if you remember the group of hard working system engineers especially when you see the announcement of massive data release.
updated. The main contents of the update are the following.
- The number of cDNA (HIT; H-Invitational transcript) entry has changed
from 175,536 to 187,156. - The number of LOCUS (HIX; H-Invitational cluster) entry has changed from
34,699 to 36,073. - The version of our mirror H-Invitational Database (H-InvDB) was upgraded
from "4.6" to "5.0".
[Databases related to the H-Invitational project]
At the following schedule, DDBJ and DAD database search is unavailable for the database update. Details are as follows:
- Date & Time: January 29, 2008(Tus) 9:00 - 18:00 (resume will be notified on this DDBJ HP)
- Suspended services:
- DDBJ and DAD search in ARSA
- From the TXSearch, link to ARSA
- From search window item DNA on the DDBJ top page, link to ARSA
- Note: During the above period, ARSA is still available except for the above 2 databases search.
If you would like to hasten to the search of the above 2 databases, please substitute SRS. - Alteration:In the DDBJ database in the ARSA, amino acid translation in the CDS feature was excluded from the specifiable features from this release72.
In the ARSA, nucleotide sequense data is also not included in the query values.
Thank you for your understanding and cooperation.
Maintenance works finished. Thank you for your cooperation.(Jan. 29, 2008 at 18:00)
and the total number of residues reached 2,995,558,433.
FTP site for DB download
because of the emergency network maintenance.
All DDBJ network services and NIG supercomputer (supernig) service will also be unavailable.
Date & Time:January 22 (Tue), 2008 from 15:00 to 16:00 (JST)
Thank you very much for your understanding and cooperation.
Maintenance works finished, and the services are resumed. Thank you for your cooperation.(21:00)
"DDBJ with new system and face"
H. Sugawara, O. Ogasawara, K. Okubo, T. Gojobori and Y. Tateno
Nucleic Acids Research, 2008, Vol. 36, Database issue D22-D24
(Summary) DDBJ activitiew of 2007 (Data submissions and release, New Computer System, ARSA(New keyword search system) enhancement, etc).
DDBJ network service and NIG supercomputer (supernig) service will also be unavailable.
- Date:January 16(W), 2008 at 12:00 - 13:00 JST
10 minutes interception during the above time
Thank you very much for your understanding and cooperation.
- What is RSS?
- RSS is a shortened name of Rich Site Summary or Really Simple Syndication, and a number of web site has used to push the information on headline, outline, update date, the link to the web site and so on to the user.
- How to read RSS ?
- You can subscribe to RSS of any Web site by Google, Yahoo! and other portal sites, if you are a registered user of the portal site. Please find the example of subscription in the following in the case of Goole:
- Why you should use RSS?
- You need not visit the site every morning to find what's new. You will be notified by the site that the site is updated. In the case of GIB RSS, the list of new genomes and updated genomes is delivered to you. You click the genome of you interest in the list and you are directly guided to the web page of the genome that you choose.
- How to register to the GIB RSS
- You may want to install a program of RSS reader by yourself, instead of using Google, Yahoo! or other portal sites. We explain the usage of RSS program installed in Windows machine:
- Install a RSS reader. The following figure is RssReader
- Register RSS delivery site of GIB(http://gib.genes.nig.ac.jp/rss.php) to the installed RSS reader.
- How to register to the GIB RSS
- You may want to install a program of RSS reader by yourself, instead of using Google, Yahoo! or other portal sites. We explain the usage of RSS program installed in Windows machine:
- Install a RSS reader. The following figure is RssReader(http://www.rssreader.com/)。
- Register RSS delivery site of GIB(http://gib.genes.nig.ac.jp/rss.php) to the installed RSS reader.
- The line of GIB is added.
- Select the GIB line (the box 3 in the figure) and the list from GIB is displayed.
- Choose one of the genomes in the list (the box 4 in the figure) and you will know if it is updated or newly added.
Please forward your request to this type of alert system and we will expand RSS to other DDBJ services
The periodical release and the new data are available by FTP download from the "FTP/Web API" page.
As was already notified, DDBJ proceeded deletion of the both phone and fax numbers, and E-mail address from the flat files of the entries submitted to DDBJ. The retrofit of the all entries was completed by the issue of this release 72. Hereafter, database users become difficult to contact submitters based on the information described in the DDBJ flat files. When you wish to contact to the submitter(s) of an entry of your interest, please contact us using the prescribed inquiry form placed in the DDBJ HP.