Patent column from DDBJ
Quote from Mail Magazine : No.62&63 (Sep.02,2011) ~ No.71&72
(Jul.26,2012) and April 2013 Issue.
*Please note about different of last information.
Hideo Aono (DDBJ Patent Annotator)
1. Patent data distribution for Japan Patent Office (JPO)
1. Introduction
In this column, I would like to introduce about patent data exchange in the International Nucleotide Sequence Databases (INSD) and Japan Patent Office (JPO) data.
2. Data distribution from INSD
DDBJ is constructing INSD with EMBL-Bank in EBI (European Bioinformatics
Institute) and GenBank in NCBI (National Center for Biotechnology
Information).
INSD distributes not only scientific data but also patent data
transferring from each patent office as follows.
- Japan: Japan patent office (JPO) → DDBJ
- South Korea: Korean Intellectual Property Office (KIPO) → DDBJ
- USA: United States Patent and Trademark Office (USPTO) → GenBank
- Europe: European Patent Office (EPO) → EMBL-Bank
Patent data contains nucleotide sequence and amino acid sequence. After
distributed nucleotide sequence data, it was exchanged among INSD.
However, INSD does not exchanged amino acid data.
DDBJ distributes JPO and KIPO amino acid data on DDBJ anonymous FTP
site.
DDBJ anonymous FTP:
JPO file: jpo_ddbj_aa*.seq.gz (*=file number)
KIPO file: kipo_ddbj_aa.seq.gz
3. Patent data summary for JPO
3-1) JPO data
DDBJ distributes nucleotide sequence and amino acid data containing publication of unexamined patent applications. Therefore, new distribution sequences do not have the patent right. When you would like to confirm the progression status of patent examination, you need checking the registration status using by retrieval site for (Industrial Property Digital Library) in Japan.
3-2) Kinds of JPO patent data and distribution frequency
DDBJ receives three kind of patent data from JPO;
- Publication of patent applications
- International application published under the Patent Cooperation Treaty (PCT)
- Japanese translations of PCT international publication for patent applications
DDBJ distributes the regular data at once a month and infrequently irregular data in some cases.
3-3) Patent data contents
JPO data of DDBJ flat file (FF) has enough information such as the front page of patent publication by the following contents; sequence, organism name, invention title, applicant name(s), inventor name(s), publication number, publication date, application number, application date, priority date, comment and sequence’s feature information.
Example: DL000001
3-4) Patent applied sequence submission to DDBJ
If you have sequence(s) for patent application, you do not have to submit to DDBJ. Because JPO transfers those sequences to DDBJ and your sequence(s) will be opened to the public through DDBJ. However, if you need the accession number to submit a paper before apply for a patent, you should care since sequence becomes public knowledge in case of released from DDBJ. In this case, please confirm to JPO before your submission.
4. Author comments
I hope to help understanding about JPO patent data. I will introduce about KIPO data and retrieval method of patent sequence in KIPO related organization at next column.
2. Patent data distribution for Korean Intellectual Property Office (KIPO)
1. Introduction
In previous column, I introduced about patent
data for Japan Patent Office (JPO). In this column, I would like to
introduce about patent data of Korean Intellectual Property Office
(KIPO) and data search service in DDBJ.
DDBJ has just distributed some of KIPO data. Therefore, I would like to
introduce also search service of patent applied sequence at Korean
Bioinformation Center (KOBIC) and Korea Intellectual Property Rights
Information Service (KIPRIS) as KIPO related organization.
2. Summary of KIPO data
DDBJ has provided the nucleotide sequence and amino acid sequence containing publication of unexamined patent applications from KIPO since March 2008.
KIPO data is transferred to DDBJ as follows;
- KIPO sends the patent sequence data to KOBIC.
- By cooperation of Dr. Byungwook Lee in KOBIC, KIPO submission files are made by him and transferred from KOBIC to DDBJ.
- DDBJ exchanges the data format and distributes KIPO data.
DDBJ distributed the following 282,161 entries assigned the prefix “DI”
of KIPO accession number, until now.
KIPO data submission is suspended by the problem of submission format
from April 2009. Therefore DDBJ is discussing about efficient method for
data exchange with KOBIC and KIPO.
Nucleotide sequence data : DI000001 - DI168562 (168,562 entries)
Amino acid sequence data : DI500001 - DI613599 (113,599 entries)
Please see Data example of Fig.1 and Fig.2.
3. Data search for KIPO data
DDBJ
DDBJ provides the search service for KIPO nucleotide and amino acid sequence (Table.1).
| getentry | ARSA | BLAST | File ftp service | |
|---|---|---|---|---|
| Nucleotide sequence | ○ | ○ | ○ | PAT division files (*1) in DDBJ release |
| Amino acid sequence | ○ | × | ○ | Cumulated file on DDBJ anonymous site (*2) |
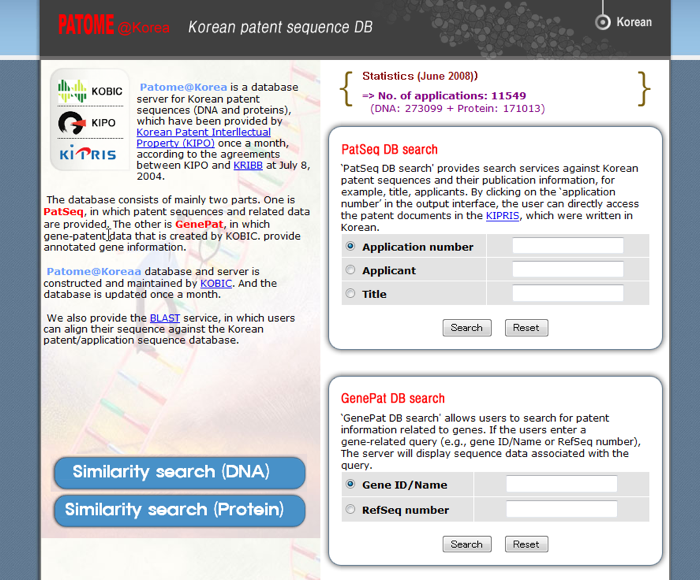
Korean Bioinformation Center (KOBIC)
PATOME@Korea is Koran patent database for nucleotide and amino acid sequence in KOBIC. “PatSeq DB search” can search by application number, applicant name, invention title in English. In addition, PATOME@Korea provides the similarity search for DNA and protein against patent applied sequences in Korea (Please see Fig.3).
Korea Intellectual Property Rights Information Service (KIPRIS)
KIPRIS is the search system of Korean patent publication. PATOME@Korea can search the patent publication in Korean. In contrast, KPA (Korean Patent Abstracts) search of KIPRIS can refer the front page and abstract of patent publication in English retrieved by keyword, application number, publication number, applicant name and inventor name (Please see Fig.4). K2E-PAT (Korean to English Patent Automatic Translation) has pay service for the provision of full text by English machine translation.
Author comments
KIPO suspends their submission to DDBJ. We will restart KIPO submission
as soon as possible.
At next column, I will introduce about the structure of DDBJ Flat Files
(FF) for JPO patent data.
3. Flat File structure for Japan Patent Office (JPO) -First part-
1. Introduction
I introduced about patent data of Japan Patent Office (JPO) in first column and Korean Intellectual Property Office (KIPO) in second column. In this time, I would like to explain about Flat File (FF) structure for JPO nucleotide and amino acid sequence data as two parts. I will introduce about JPO data property, summary of FF structure and relationship between patent application contents and description part on FF in first part.
2. Property of JPO data FF
JPO data FF has fundamental application information of patent publication compared with FF for United States Patent and Trademark Office (USPTO) and European Patent Office (EPO). Because JPO makes the submission file (Fig. 1) containing application information based on WIPO (World Intellectual Property Organization) ST 25 format submitted from applicants. Thereafter, DDBJ makes FF reflected almost contents of JPO submission file (Fig. 2).
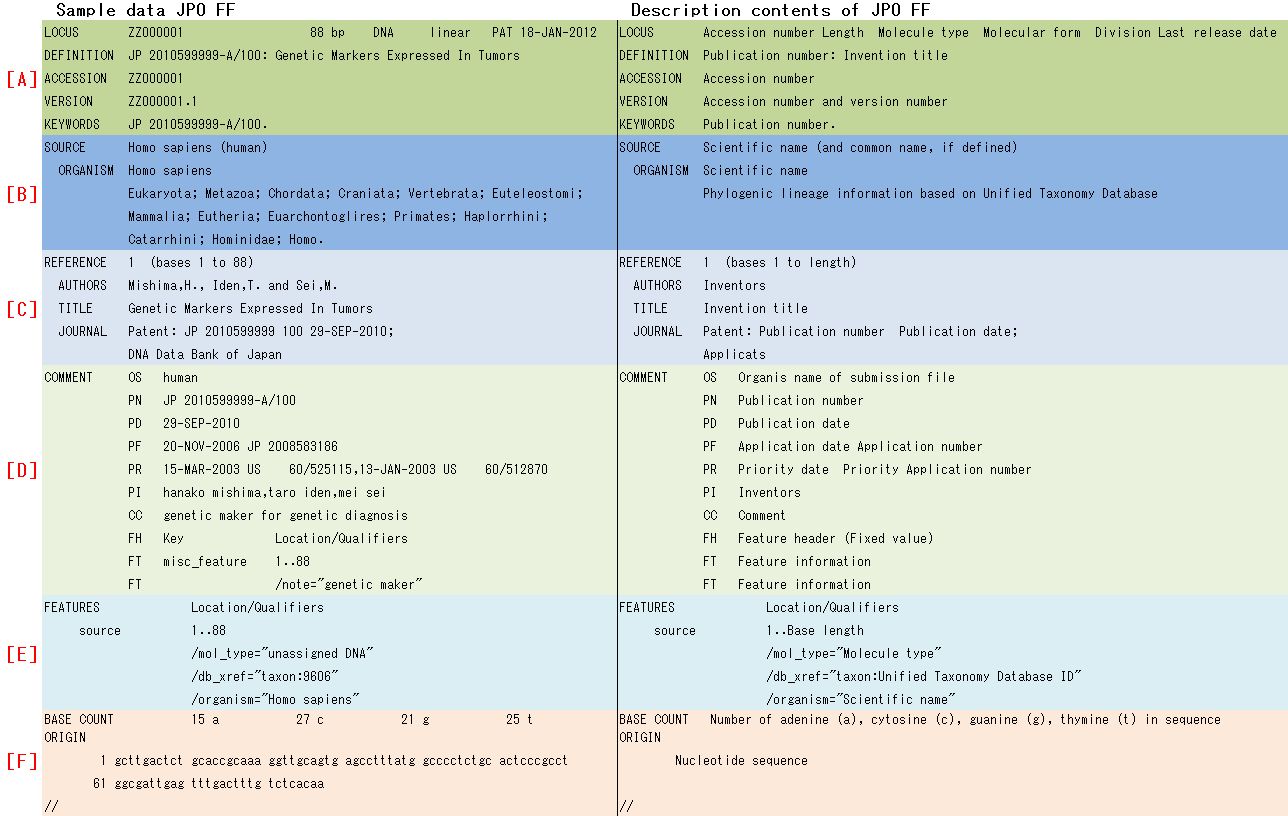
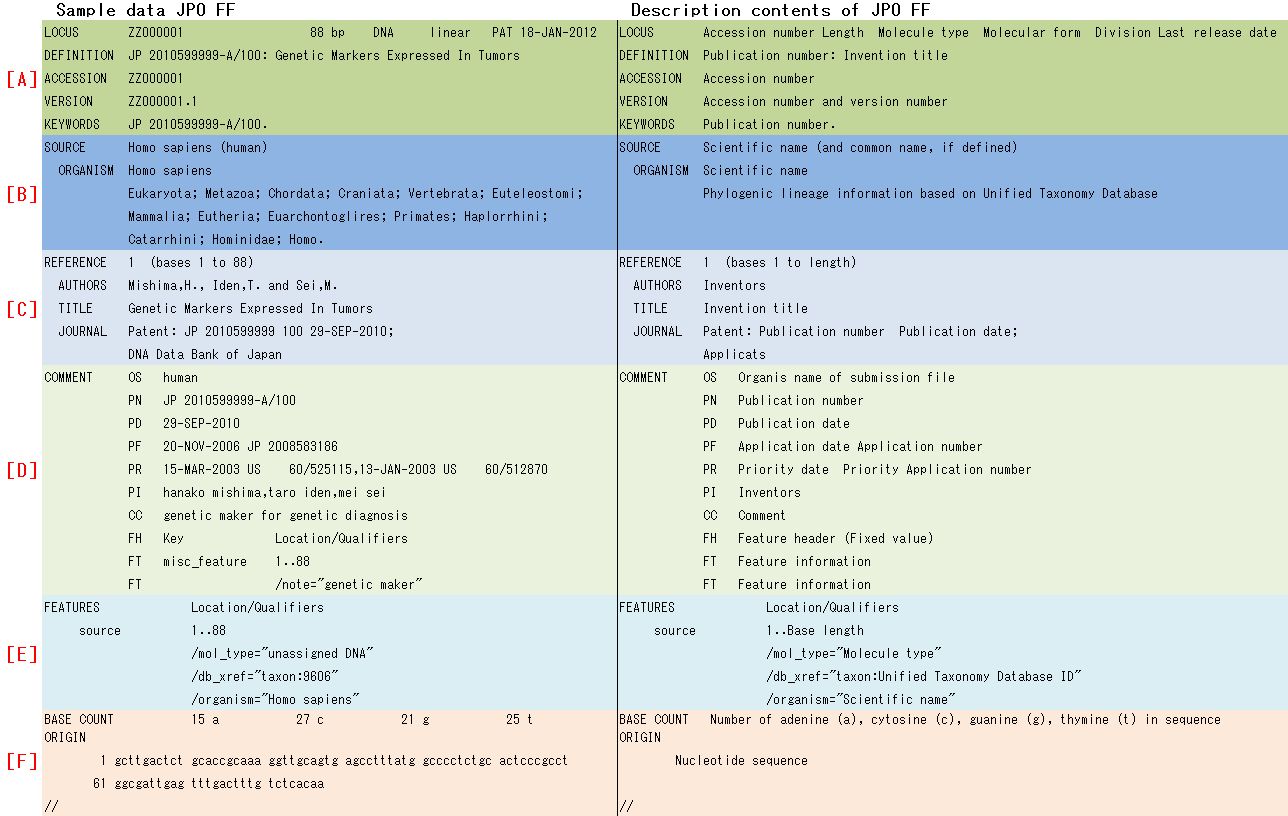
3. FF structure and description contents
I showed FF structure for nucleotide sequence data dividing by six parts (LOCUS Block, SOURCE Block, REFERENCE Block, COMMENT Block, Feature Block and Sequence Block) as different color with sample data and description contents in Fig. 2.
Each Block name of symbol alphabet is as follows;
[A] LOCUS Block, [B] SOURCE Block, [C] REFERENCE Block, [D]
COMMENT Block, [E] Feature Block, [F] Sequence Block
4. Patent application contents on FF
I showed correspondence relationship between patent application contents and description parts on JPO FF (Table. 1).
| hContents of patent application | Describing line on JPO FF |
|---|---|
| Publication number | DEFINITION line [A], KEYWORDS line [A], JOURNAL line [C], PN line [D] |
| Publication date | JOURNAL line[C] , PD line[D] |
| Application number | PF line [D] |
| Application date | PF line [D] |
| Applicants | JOURNAL line [C] |
| Inventors | AUTHORS line [C], PI line [D] |
| Invention title | DEFINITION line [A], TITLE line [C] |
| Priority application number | PR line [D] |
| Priority date | PR line [D] |
| Feature information | FT line [D] |
| Organism name (JPO submission file) | OS line [D] |
| Organism name (converted name in DDBJ) | SOURCE line [B], ORGANISM line [B], /organism qualifier [E] |
Table. 1 Application contents and description parts on JPO FF
* Alphabet symbol after line name is corresponding to FF Block part in Fig. 2.
Patent application information is described mainly at [C] REFERENCE
Block and [D] COMMENT Block. Amino acid sequence data is also
described on same Blocks. Invention title is set on both DEFINITION line
of [A] LOCUS Bloc and TITLE line of [C] REFERENCE Block.
In specially, applicants name is only set JOURNAL line on [C]
REFERENCE Block (Fig. 3).

5. Author comments
At second part, I will explain each line description of six Blocks in detail. Moreover, I would like to introduce the conversion process of organism name from JPO submission file in DDBJ.
4. Flat File structure for Japan Patent Office (JPO) -Second part-
1. Introduction
I described about summary of Flat File (FF) structure of Japan Patent Office (JPO) in previous column at first part. In second part, I will introduce FF structure in more detail, conversion process of organism name from JPO submission file and the format of Patent publication number.
2. FF structure and description contents
I showed FF structure for nucleotide sequence data dividing by six parts ([A] LOCUS Block, [B] SOURCE Block, [C] REFERENCE Block, [D] COMMENT Block, [E] Feature Block and [F] Sequence Block) as different color with sample data and description contents in Fig. 1.
2-1) [A] LOCUS Block
LOCUS Block has LOCUS, DEFINITION, ACCESSION, VERSION and KEYWORDS line (Table. 1).
| Line name | Description Contents (NA: Nucleotide sequence data. AA: Amino acid sequence data) |
|---|---|
| LOCUS [Example] NA: Fig.1 AA: Fig.2 |
Accession number Sequence length number (NA: bp, AA: aa) Molecule type (NA: DNA or RNA, AA: PRT) Molecular form (NA: linear, AA: not described) Division (PAT) Last release date (If the entry is updated and reopened to public site, this date will be changed.) |
| DEFINITION | Publication number and Invention tile (Same as title line of REFERENCE Block) Example:JP 2010599999-A/100: Genetic Markers Expressed In Tumors |
| ACCESSION | Accession number The accession number prefixes of JPO data are as follows; NA: E, BD, DD, DJ, DL, DM, FU, FV, FW, FZ, GB, HV AA: E, BD, DD The accession number prefix of KIPO data is as follows; NA: DI AA: DI |
| VERSION | NA: Sequence version number with Accession number Example: ZZ000001.1 The data open to public for the first time is version number as “1”. If sequence is updated, version number is increased. AA: Not set on VERSION line |
| KEYWORDS | Patent publication number JP header: Number of Publication of patent applications in Japan and Japanese translations of PCT international publication for patent applications. WO header: Number of International application published under the Patent Cooperation Treaty (PCT) Please also refer their number format in section (3). |
Table. 1: Description Contents of LOCUS Block
In case of amino acid sequence data, LOCUS line has the different output format (Fig.2). Its line has Accession number, Sequence length number, Molecule type, Division and Last release date.
2-2) [B] SOURCE Block
Scientific name on SOURCE and ORGANISM lines is converted from NCBI Taxonomy Database (Table. 2). Please also refer at section (2).
| Line name | Description Contents |
|---|---|
| SOURCE | Scientific name (Common name) SOURCE line is set scientific name and common name. Scientific name is converted from organism name on OS line of COMMENT Block based on NCBI Taxonomy Database. Moreover, if scientific name has common name (example: human) in NCBI Taxonomy Database, common name is set after scientific name. [Example] SOURCE Homo sapiens (human) |
| ORGANISM | Scientific name on fist line and its lineage information based on NCBI Taxonomy Database on second line. [Example] ORGANISM Homo sapiens Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Mammalia; Eutheria; Euarchontoglires; Primates; Haplorrhini; Catarrhini; Hominidae; Homo. |
Table. 2: Description Contents of SOURCE Block
2-3) [C] REFERENCE Block
REFERENCE Block has AUTHORS, TITLE and JOURNAL line (Table. 3).
| Line name | Description Contents |
|---|---|
| AUTHORS | Inventor name Inventor fill name is set PI line of COMMENT Block. |
| TITLE | Invention tile Same value is also set in DEFINITION line. |
| JOURNAL | First line is described the publication number and publication date after the fixed value “Patent:”on JOURNAL line. Applicant name is described on second line. Applicant information is only set JOURNAL line in FF. [Example] JOURNAL Patent: JP 2010599999 100 29-SEP-2010; DNA Data Bank of Japan |
Table. 3: Description Contents of REFERENCE Block
2-4)[D] COMMENT Block
| Line name | Description Contents |
|---|---|
| OS | Organism name in JPO submission file. SOURCE line, ORGANISM line, /organism and /db_xref are constructed by this organism name. |
| PN | Publication number and sequence number |
| PD | Publication date |
| PF | Application date, Application number |
| PR | Priority date, Priority application number |
| PI | Inventor name |
| CC | Comment |
| FH | Feature header (Fixed value: Key Location/Qualifiers) |
| FT | Feature information |
Table. 4: Description Contents of COMMENT Block
*Some old JPO data have PC line for description of international patent classification (IPC) code.
2-5) [E] Feature Block
JPO data has only source feature, same also Korean Intellectual Property
Office (KIPO) data (Table. 5).
Nucleic acid sequence data has /mol_type, /db_xref and /organism
qualifiers.
Amino acid sequence data does not have /db_xref qualifier.
| Qualifier name | Description Contents(NA: Nucleotide sequence data. AA: Amino acid sequence data) |
|---|---|
| /mol_type | NA: unassigned DNA,unassigned RNA AA: Not set |
| /db_xref | Taxonomy ID of NCBI Taxonomy Database setting after fixed value “ taxon:” |
| /organism | Scientific name based on NCBI Taxonomy Database |
Table. 5: Description Contents of Feature Block
(Organism information update based on NCBI Taxonomy Database)
DDBJ started adding Taxonomy ID of /db_xref to JPO and KIPO data from
May 2010. DDBJ will update the reconstruction of SOURCE line, ORGANIS
line and /organism based Taxonomy ID, once a year.
2-6) [F] Sequence Block
Nucleotide sequence data has BASE COUNT line (Fig.1) which is described
the number of adenine (a), cytosine (c), guanine (g), thymine (t).
In case of Amino acid sequence data, BASE COUNT line is not output
(Fig.3).
3. Conversion process of scientific name
3-1) Conversion of scientific name from JPO submission file
Original organism name in JPO submission file is described on OS line of COMMENT Block. Scientific name is converted from its name on OS line and set to SOURCE line, ORGANISM line and /organism qualifier based on NCBI Taxonomy database. Its lineage information is also constructed and set on ORGANISM line.
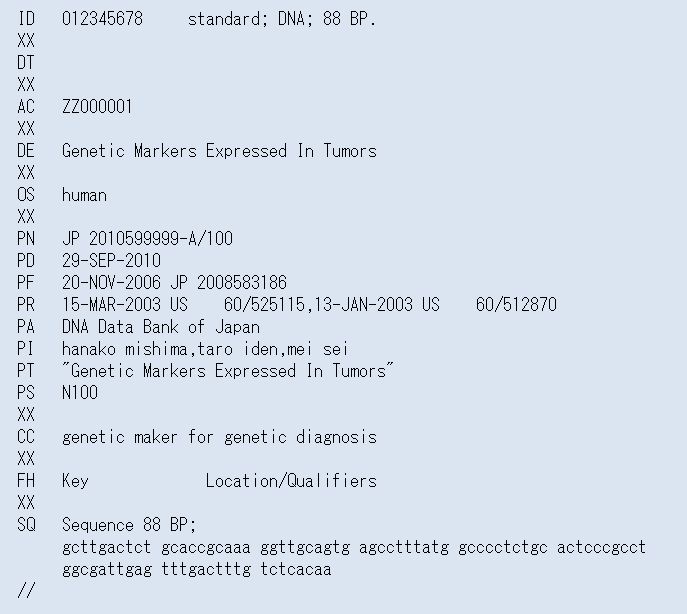
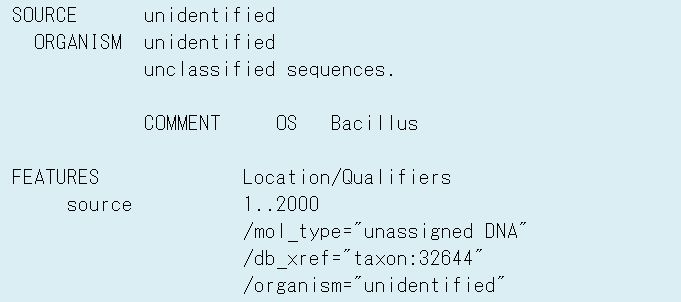
3-2) Unidentified organism name
In case of organism name described by applicants is not found in NCBI
Taxonomy database, its name is converted to “unidentified” on SOURCE
line, ORGANISM line and /organism qualifier (Fig.4).
Original organism name described by applicants is set on OS line at
COMMENT Block (Fig.1, Table 4).
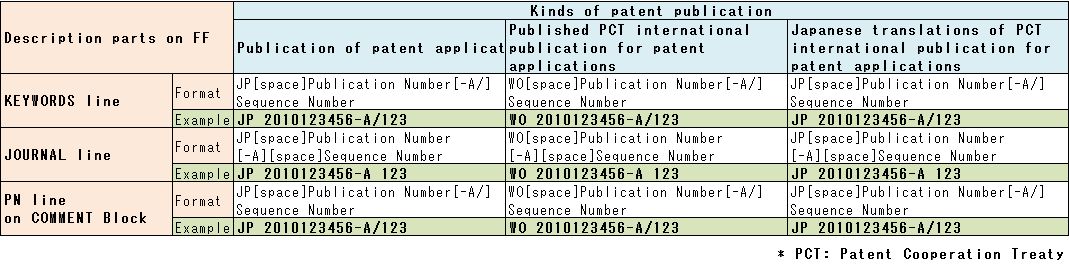
4. Format of patent publication number and Description parts on FF
4-1) Format of patent publication number
DDBJ received three kind of patent publication for [1] Publication of
patent applications, [2] Published Patent Cooperation Treaty (PCT)
international publication for patent applications and [3] Japanese
translations of PCT international publication for patent applications
(Table 6).
Publication of patent applications and Japanese translations of PCT
international publication for patent applications have same format of
publication number with “JP” in head of its number. PCT international
publication for patent applications have “WO”in head of publication
number.
4-2) Format of patent publication number
Publication number is set on KEYWORDS line of LOCUS Block, JOURNAL line of REFERENCE Block and PN line of COMMENT Block (Fig.5).
Author comments
In this time, Patent column is final. When you would like to know FF structure and patent data property for JPO and KIPO data, please refer to my columns. If I have a chance, I will explain how to search the patent data by DDBJ tools and improvement points of JPO and KIPO FF.
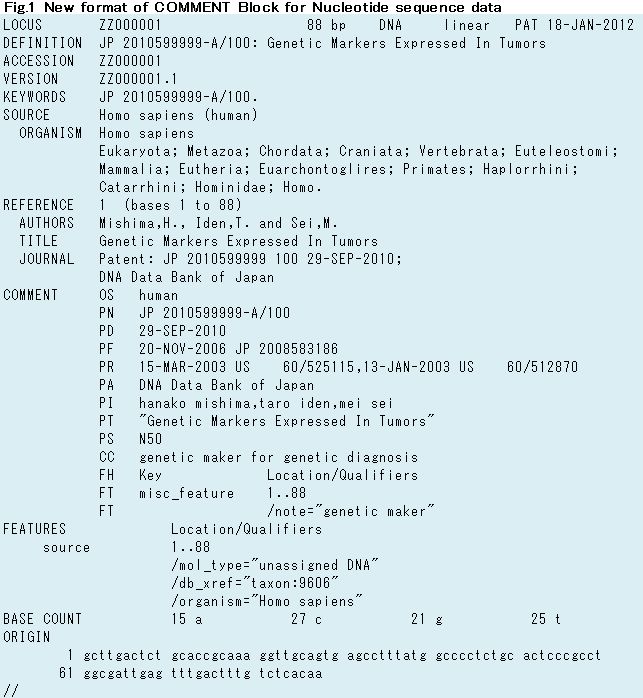
5. New COMMENT format for Japan Patent Office (JPO) data
1. Introduction
DDBJ makes an improvement of COMMENT Block on Flat File (FF) for Japan Patent Office (JPO). In this column, I would like to introduce new format of COMMENT Block in detail.
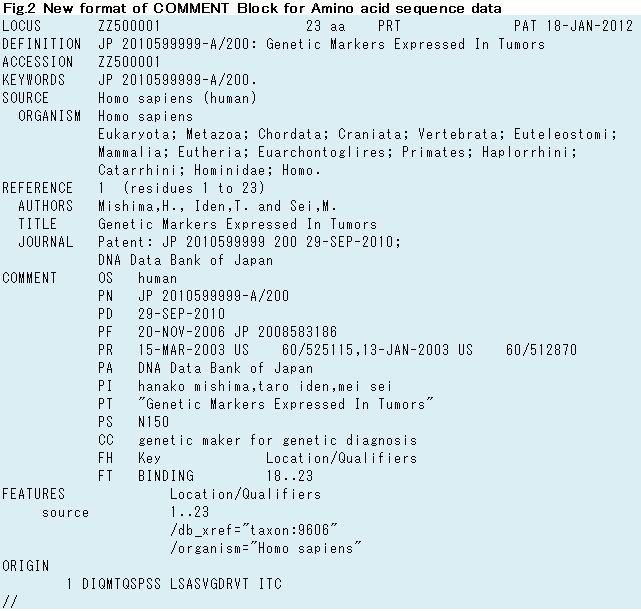
DDBJ re-constructed FF of all JPO entries to reflect new COMMENT format. New format of Nucleotide sequence data has been released since DDBJ release 90 (Fig.1) and Amino acid sequence data has been distributed JPO cumulated files on Anonymous FTP since September 2012 (Fig.2).
2. New format of COMMENT Block
JPO data has almost basic information for patent application. Therefore,
user can get its information without referring patent publication.
However, since patent information was set on different part of FF, user
had to know the FF structure to get them in advance. In specially,
applicants name is only set JOURNAL line on REFERENCE Block (Fig.3 &
Fig.4).
Therefore, DDBJ made describing basic patent information on COMMENT Block as follows (Fig.4).
COMMENT Block was added new lines of PA (Applicant name), PT (Invention title) and PS (Sequence number) in all nucleotide and amino acid sequence data (Table.1).
| hNew line nameh | Description |
|---|---|
| PA line | Applicant name JOURNAL line on REFERENCE Block is also described (Fig.3). |
| PT line | Invention title DEFINITION, TITLE line on REFERENCE Block are also described (Fig.1 & Fig.2). |
| PS line | Sequence number Sequence number for Sequence listings setting after prefix letter “N”. |
Table.1 New lines of COMMENT Block
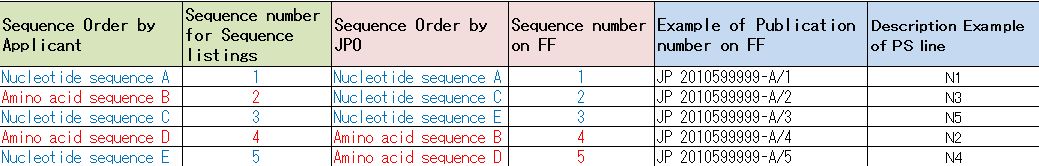
3. Sequence number
3-1:
There was mismatch problem of Sequence number between Sequence listings and JPO submission file. Old format FF was not described Sequence number of Sequence listings. Therefore, DDBJ added PS line on COMMENT Block to set its number.
3-2:
When applicants applied patent, they made Sequence listings according to
the guideline: WIPO ST.25. It was put ordering number from 1 to
nucleotide and amino acid sequences based on patent claims by
applicants.
On the other hand, JPO changes sequence order and makes nucleotide
sequence block and amino acid sequence block from Sequence listings when
they make the submission files to DDBJ. After that, they set new
Sequence number in order of nucleotide sequences, amino acid sequences
(Table.2).
3-3:
Sequence number indicated by JPO is reflected with patent publication
number on DEFINITION, KEYWORDS, JOURNAL line of REFERENCE and PN line of
COMMENT Block.
Publication of patent applications and Japanese translations of Patent
Cooperation Treaty (PCT) international publication for patent
applications have same format of publication number with “JP”. PCT
international publication for patent application has “WO” in head of
publication number (Fig.1, Fig.2 and Table.3).
| hDescription part of FFh | hFormat of patent publicationh |
|---|---|
| DEFINITION KEYWORDSPN line of COMMENT Block |
JP[space]Patent publication number[-A/]Sequence number WO[space]Patent publication number[-A/]Sequence number |
| JOURNAL line of REFERENCE Block | JP[space]Patent publication number[-A][space]Sequence number WO[space]Patent publication number[-A][space]Sequence number |
Table.3 Description part and Format of patent publication on FF
3-4:
Sequence number Sequence listings are set after prefix letter “N” on PS line. Therefore, you can retrieve appropriate data by ARSA (All-Retrieval of Sequence and Annotation) with publication number and “N” with Sequence number.
Author comments
DDBJ continues to make an improvement of Patent FF for user. As next step, we are planning FF put on direct link of the patent publication.






![Fig.4 Description of each line on REFERENCE [A] and COMMENT Block [B]](/assets/images/ddbj/5_Fig4.jpg)