MetaboBank
Metadata
MAGE-TAB
MicroArray Gene Expression Tabular (MAGE-TAB) was developed to represent functional genomics data in a structured way and has been used by ArrayExpress and GEA. MAGE-TAB is also used in proteomics fields and is becoming a standard in omics fields.
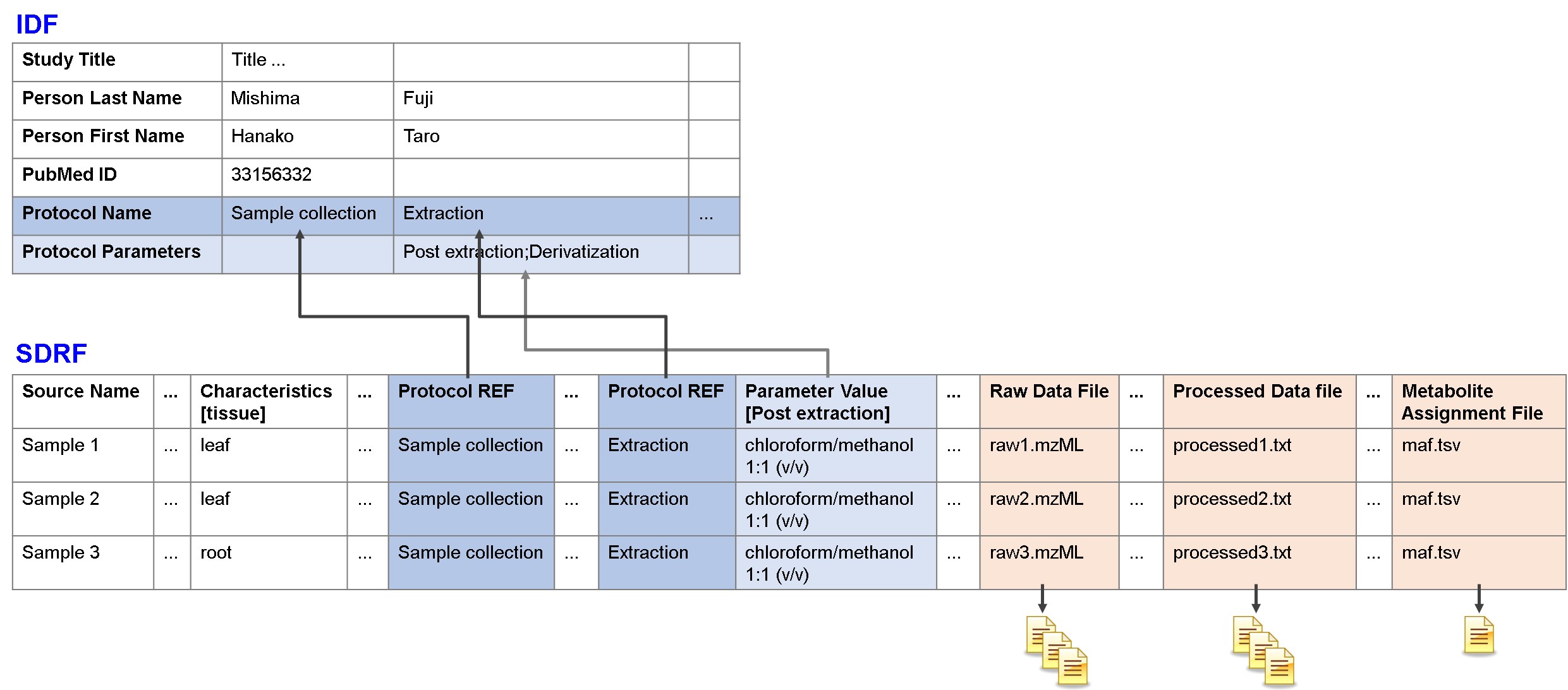
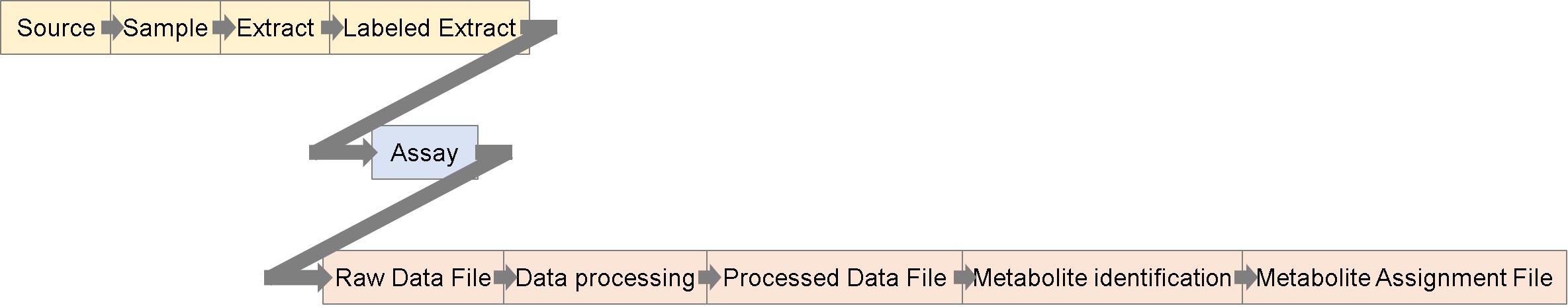
MAGE-TAB consists of two parts, IDF and SDRF. IDF describes study design, and SDRF describes sample characteristics and relationship between sample and data files. IDF and SDRF are linked by protocols. Metadata and data files are linked by SDRF.
Metadata excel
Download a metadata excel file designed for each submission type and fill in the file. Submit data of different types as separate studies.
- Mass spectrometry, chromatography
- Liquid chromatography-mass spectrometry (LC-MS, download)
- Liquid chromatography, diode array detector-mass spectrometry (LC-DAD-MS, download)
- Gas chromatography-mass spectrometry (GC-MS, download)
- Two dimensional gas chromatography-mass spectrometry (GCGC-MS, download)
- Gas chromatography, flame ionization detector-mass spectrometry (GC-FID-MS, download)
- Capillary electrophoresis-mass spectrometry (CE-MS, download)
- Mass spectrometry, direct injection
- Mass spectrometry imaging (MSI, download)
- Nuclear magnetic resonance spectroscopy (NMR, download)
IDF
IDF (Investigation Description Format) is a file describing study overview, experimental design, protocols, publication and submitter information.
IDF fields
- MAGE-TAB Version
- Version number of MAGE-TAB. Fixed to 1.1. Filled by MetaboBank.
- Comment[MetaboBank accession]
- Accession number assigned by Metabobank (e.g. MTBKS1). Filled by MetaboBank.
- Study Title
- The overall title of the study.
- Study Description
- A short paragraph describing the study as free-text. The text should clearly explain what you did in your study. In this field, ASCII, Greek characters and symbols [° μ ± ≠ ≒ < > ← ↑ ↓ → ↔ Å] are allowed for richer description.
- Experimental Design
- The experiment design types which are applicable to this study. These terms should come from controlled terms.
- Experimental Factor Name
- A user-defined name for each experimental factor studied by the experiment. These experimental factors represent the variables (parameters) within the investigation. The actual values of these variables will be listed in the SDRF file, in “Factor value[<factor name>]” columns. For example, an experiment studying the effect of different temperature (heat stress) on a cell culture would have “temperature” as an experimental variable with “Unit” column to indicate the unit in the accompanying SDRF Factor Value[] columns (e.g. Factor Value[temperature]).
- Experimental Factor Type
- A term describing the type of each experimental factor. Filled by MetaboBank.
- Person Last Name
- The last name of each submitter. Enter last names of submitters in each column. Submitters have edit rights to the submission and correspondence will be directed to the submitter. They will also be the main contact for once the study is public. Use Comment[Contributor] to list contributors.
- Person First Name
- The first name of each submitter. Enter first names of submitters in each column.
- Person Mid Initials
- The middle name of each submitter. Enter middle names of submitters in each column.
- Person Email
- The Email address of each submitter. Enter addresses of submitters in each column. The Email address is not publicly displayed.
- Person Affiliation
- The organization affiliation of each submitter. Enter affiliations of submitters in each column.
- Person Roles
- The role(s) performed by each person. Only “submitter” role is permitted. Filled by MetaboBank.
- PubMed ID
- The PubMed IDs of the publication(s) associated with this study (where available).
- Publication DOI
- A Digital Object Identifier (DOI) for each publication (where available). When PubMed ID and DOI are available, use PubMed ID.
- Public Release Date
- The date the submission was initially released publicly. Filled by MetaboBank.
- Term Source Name
- The names of the Term Sources (ontologies or databases) used within the IDF and SDRF. The “Term Sources” are defined in the IDF and may be used throughout the IDF and SDRF. This name will be used in all corresponding “Term Source REF” fields.
- Term Source File
- A filename or valid URI at which the Term Source may be accessed.
- Term Source Version
- The version of the Term Source used throughout the IDF and SDRF.
- SDRF File
- The name of the SDRF file accompanying this IDF file. Filled by MetaboBank.
- Comment[Study type]
- The study types which are applicable to this study (e.g. targeted metabolite profiling, lipid profiling). These terms should come from controlled terms.
- Comment[Experiment type]
- The experiment types which are applicable to this study (e.g. liquid chromatography-mass spectrometry, capillary electrophoresis-mass spectrometry). These terms should come from controlled terms. More than one type can be added. Fixed terms for the submission type are filled by MetaboBank.
- Comment[Submission type]
- The type of this submission (e.g. LC-MS, GC-MS). These terms should come from controlled terms. Filled by MetaboBank.
- Comment[BioProject]
- A related BioProject accession (for example, PRJDB1).
- Comment[Related study]
- Related accession number(s) of MetaboBank (MB) or other databases. List accessions in the format of “DB:ID” in tab-delimited fields. Example, MB:MTBKS202<tab>MB:MTBKS203<tab>Metabolonote:SE112.
- Comment[Contributor]
- Name of each contributor. Contributors such as technical staffs can be listed in the Comment[Contributor] regardless of they are submitters or not. The contributors do not automatically have edit rights if they are not specified as submitters.
Example, Mishima Naoko, Fuji San, Shizuoka Ken. - Comment[Submission Date]
- The date this data was submitted. Filled by MetaboBank.
- Comment[Last Update Date]
- The date of last update. Filled by MetaboBank.
IDF Protocols
Protocols are central for reproducibility purposes and should provide a detailed description of the steps taken in the study.
- Protocol Name
- The names of the protocols used. Use the protocol type for the name when there is one protocol for the type. If there are more than one protocols for the type, differentiate them by adding 1, 2, … (e.g. Data transformation 1, Data transformation 2).
- Protocol Type
- The type of the protocol. Required protocols are different for each submission type.
| Protocol type | Description | Submission type |
|---|---|---|
| Sample collection | Describe the origin of samples, any relevant treatment, time points etc and the collection and storage procedure. | All types |
| Extraction | Describe any extraction or preparation methods applied to the sample before analysis. Please also include any control samples prepared for the assay, e.g. pooled samples, standards, quality control, solvent blank etc | Other than MSI |
| Chromatography | Provide details of the instrument and column used (manufacturer), mobile phase and gradient, and settings such as temperatures, flow rate, injection volume. | LC-MS,LC-DAD-MS,GC-MS,GCGC-MS,GC-FID-MS |
| Mass spectrometry | Provide details of the instrument used (manufacturer), ion source, ionisation mode (positive/negative), m/z range, and specific parameters such as temperatures, voltages, flow rates, scan rates. | Other than NMR |
| Data processing | Provide details of methods/pipelines and software used to transform the raw data. | All types |
| Metabolite identification | Provide details of methods/pipelines, reference databases and software used to identify features and/or annotate metabolites. | All types |
| Capillary Electrophoresis | Provide details of the instrument and column used (manufacturer), mobile phase and gradient, and settings. | CE-MS |
| Direct infusion | Provide details of the direct infusion methods. | DI-MS |
| Flow injection analysis | Provide details of the flow injection analysis methods. | FIA-MS |
| Preparation | Describe sample preparations such as mounting, preservation, tissue modification, sectioning and matrix. | MSI |
| Histology | Describe histological details such as stain. | MSI |
| NMR sample | Describe NMR samples such as tube type, solvent, sample pH and temperature. | NMR |
| NMR spectroscopy | Provide details of NMR instrument, probe and magnetic field strength etc | NMR |
| NMR assay | Provide details of NMR assay. | NMR |
- Protocol Description
- A free-text description of the protocol. This text should be included in a single tab-delimited field. In this field, ASCII, Greek characters and symbols [° μ ± ≠ ≒ < > ← ↑ ↓ → ↔ Å] are allowed for richer description.
- Protocol Parameters
- A semicolon-delimited list of parameter names. Required parameters are different for each submission type. See SDRF Protocol Parameters for details.
- Protocol Hardware
- The protocol hardware is the instrument that was used to capture the sample. If multiple instruments are used, they should be separated by semicolon (;). In this field, ASCII, Greek characters and symbols [° μ ± ≠ ≒ < > ← ↑ ↓ → ↔ Å] are allowed for richer description.
- Protocol Software
- The software used by the protocol.
SDRF
SDRF (Sample and Data Relationship Format) is a file describing sample characteristics and relationship between samples, measurement instruments and data files. SDRF is a table represents omics experimental flow, starts from source samples and ends with data files.
SDRF columns
- Source Name
- A unique identifier from a particular source. Use a sample name of BioSample in most cases.
- Characteristics
- Sample attributes. Use BioSample attributes used to describe sample characteristics (e.g. organism, strain). The sample_title, description and BioSample accessions are entered in the Comment columns of Source Name. The other non-sample characteristics attributes such as bioproject_id and locus_tag_prefix are excluded from SDRF.
- Protocol REF
- The column for referencing a protocol defined in IDF by its name. This is the column marking the start of data pertaining to the referencing protocol. The protocol name must be present in all rows of this column.
- Sample Name
- A unique identifier from a particular sample. Use a sample name of BioSample in most cases.
- Extract Name
- A unique identifier from a particular extracted material.
- Labeled Extract Name
- A unique identifier from a particular extract chemically labeled by isotopes. Optional for non-labeled samples. Leave blank if you don’t have one. See FAQ: How to describe samples labeled by isotopes? for details.
- Label
- If you used a chemical or biochemical marker in the sample such as a radioactive isotope which is bound to a material in order to make it detectable in an analytical instrument then enter it here. Leave blank if you don’t have one. See FAQ: How to describe samples labeled by isotopes? for details.
- Assay Name
- A unique identifier from a particular assay name. Technical replicates are represented by rows having same sample names and different assay names with technical replicate comments.
- Comment[technical_replicate]
- Technical replicates such as 1, 2 and 3.
- Raw Data File
- The column to enter raw (unprocessed) data files. If your data has been processed into one of the open-source raw data formats (e.g. mzML and nmrML etc), then add them here.
Files can be specified in several ways.
Enter each filename for each sample in single column.
| Raw Data File | Comment[Raw Data File md5] |
|---|---|
| sample1.RAW.gz | … |
| sample2.RAW.gz | … |
Enter each tar/zip archived filename for each sample in single column.
| Raw Data File | Comment[Raw Data File md5] |
|---|---|
| sample1.RAW.tar.gz | … |
| sample1.RAW.tar.gz | … |
Enter each subdirectory name containing files for each sample in single column.
| Raw Data File | Comment[Raw Data File md5] |
|---|---|
| sample1/ | … |
| sample2/ | … |
Enter two filenames for a sample in two columns.
| Raw Data File | Comment[Raw Data File md5] | Raw Data File | Comment[Raw Data File md5] |
|---|---|---|---|
| sample1.RAW.gz | … | sample1.mzML | … |
| sample2.RAW.gz | … | sample2.mzML | … |
- Comment[Raw Data File md5]
- Enter MD5 hash value of raw data file here.
- Processed Data File
- The column to enter processed data files. The processed data file has a broad meaning which ranges from processed raw data files to summary table. Files can be specified in several ways.
Enter each filename for each sample in single column.
| Processed Data File | Comment[Processed Data File md5] |
|---|---|
| sample1.tsv | … |
| sample2.tsv | … |
Enter each tar/zip archived filename for each sample in single column.
| Processed Data File | Comment[Processed Data File md5] |
|---|---|
| sample1.tsv.tar.gz | … |
| sample1.tsv.tar.gz | … |
Enter each subdirectory name containing files for each sample in single column.
| Processed Data File | Comment[Processed Data File md5] |
|---|---|
| sample1/ | … |
| sample2/ | … |
Enter two filenames for a sample in two columns.
| Processed Data File | Comment[Processed Data File md5] | Processed Data File | Comment[Processed Data File md5] |
|---|---|---|---|
| sample1.tsv | … | sample1.xlsx | … |
| sample2.tsv | … | sample2.xlsx | … |
- Comment[Processed Data File md5]
- Enter MD5 hash value of processed data file here.
- Metabolite Assignment File
- A TSV file containing information about the metabolites investigated in the study. Information regarding database accession IDs, where in the spectra the metabolite is found and data pertaining to its abundance within the study samples should be reported in this file format. See Metabolite assignment file for details.
- Comment[Metabolite Assignment File md5]
- Enter MD5 hash value of metabolite assignment file here.
- Comment[maf_value_unit]
- Value unit used for the experimental data in metabolite assignment file (e.g. peak area, pico mole etc).
- Factor Value[]
- The factor values for an experiment are the values of the variables (parameters) under investigation. For example, an experiment studying the effect of different temperature (heat stress) on a cell culture would have “temperature” as an experimental variable with “Unit” column to indicate the unit.
| Factor Value[temperature] | Unit[temperature] |
|---|---|
| 37 | degree_C |
| 40 | degree_C |
- Unit[<unit category>]
- Used as an attribute column following Characteristics, Factor Value or Parameter Value. This column contains terms describing the unit(s) to be applied to the values in the preceding column. The type of unit is included in the column heading, e.g. “Unit[temperature]”.
- Image Data File
- Data files obtained in imaging experiments. The open-source data format files imzML and ibd are recommended to be included. Also submit tissue image files (png, jpg).
- Comment[Image Data File md5]
- Enter MD5 hash value of image data file here.
- Acquisition Parameter Data File
- These should contain the acquisition parameter data. In the Bruker raw data file structure, the file is called ‘acqus.txt’. Example, acqus1.txt.
- Comment[Acquisition Parameter Data File md5]
- Enter MD5 hash value of acquisition parameter data file here.
- Free Induction Decay Data File
- These should contain the free induction decay data file.
- Comment[Free Induction Decay Data File md5]
- Enter MD5 hash value of free induction decay data file here.
SDRF Protocol Parameters
Protocol Parameters supplement protocols described in IDF. The necessary and recommended parameters are different for each Submission and Protocol type.
| Protocol parameter | Submission type | Protocol type |
|---|---|---|
| Post extraction | Other than MSI,NMR | Extraction |
| Derivatization | Other than MSI,NMR | Extraction |
| Chromatography instrument | LC-MS,LC-DAD-MS,GC-MS,GCGC-MS,GC-FID-MS | Chromatography |
| Autosampler model | LC-MS,LC-DAD-MS,GC-MS,GCGC-MS,GC-FID-MS | Chromatography |
| Column model | LC-MS,LC-DAD-MS,GC-MS,GC-FID-MS | Chromatography |
| Column type | LC-MS,LC-DAD-MS,GC-MS,GC-FID-MS | Chromatography |
| Guard column | LC-MS,LC-DAD-MS,GC-MS,GCGC-MS,GC-FID-MS | Chromatography |
| Column model 1 | GCGC-MS | Chromatography |
| Column type 1 | GCGC-MS | Chromatography |
| Column model 2 | GCGC-MS | Chromatography |
| Column type 2 | GCGC-MS | Chromatography |
| Detector | LC-DAD-MS,GC-FID-MS | Chromatography |
| Signal range | LC-DAD-MS | Chromatography |
| Resolution | LC-DAD-MS | Chromatography |
| Temperature | LC-MS,LC-DAD-MS,GC-MS,GC-FID-MS | Chromatography |
| Scan polarity | All MS types | Mass spectrometry |
| Scan m/z range | All MS types | Mass spectrometry |
| Instrument | All MS types | Mass spectrometry |
| Ion source | All MS types | Mass spectrometry |
| Mass analyzer | All MS types | Mass spectrometry |
| CE instrument | CE-MS | Capillary Electrophoresis |
| Autosampler model | CE-MS | Capillary Electrophoresis |
| Column model | CE-MS | Capillary Electrophoresis |
| Column type | CE-MS | Capillary Electrophoresis |
| DI instrument | DI-MS | Direct infusion |
| FIA instrument | FIA-MS | Flow injection analysis |
| Instrument manufacturer | MSI | Mass spectrometry |
| Solvent | MSI | Mass spectrometry |
| Target material | MSI | Mass spectrometry |
| Spatial resolution | MSI | Mass spectrometry |
| Pixel size x | MSI | Mass spectrometry |
| Pixel size y | MSI | Mass spectrometry |
| Max count of pixel x | MSI | Mass spectrometry |
| Max count of pixel y | MSI | Mass spectrometry |
| Max dimension x | MSI | Mass spectrometry |
| Max dimension y | MSI | Mass spectrometry |
| Inlet type | MSI | Mass spectrometry |
| Detector | MSI | Mass spectrometry |
| Detector mode | MSI | Mass spectrometry |
| Resolving power | MSI | Mass spectrometry |
| Resolving power m/z | MSI | Mass spectrometry |
| Native spectrum identifier format | MSI | Mass spectrometry |
| Data file content | MSI | Mass spectrometry |
| Spectrum representation | MSI | Mass spectrometry |
| Raw data file format | MSI | Mass spectrometry |
| Instrument software | MSI | Mass spectrometry |
| Instrument software version | MSI | Mass spectrometry |
| Line scan direction | MSI | Mass spectrometry |
| Line scan sequence | MSI | Mass spectrometry |
| Scan pattern | MSI | Mass spectrometry |
| Scan type | MSI | Mass spectrometry |
| Number of scans | MSI | Mass spectrometry |
| Sample mounting | MSI | Preparation |
| Sample preservation | MSI | Preparation |
| Tissue modification | MSI | Preparation |
| Sectioning instrument | MSI | Preparation |
| Section thickness | MSI | Preparation |
| Matrix | MSI | Preparation |
| Matrix application | MSI | Preparation |
| Stain | MSI | Histology |
| Data processing software | MSI | Data processing |
| Data processing software version | MSI | Data processing |
| Extraction method | NMR | Extraction |
| NMR tube type | NMR | NMR sample |
| Solvent | NMR | NMR sample |
| Sample pH | NMR | NMR sample |
| Temperature | NMR | NMR sample |
| Instrument | NMR | NMR spectroscopy |
| NMR probe | NMR | NMR spectroscopy |
| Number of transients | NMR | NMR spectroscopy |
| Pulse sequence name | NMR | NMR spectroscopy |
| Magnetic field strength | NMR | NMR spectroscopy |
- Parameter Value[Post extraction]
- This column describes how the sample was extracted into a solvent prior to being injected into the analytical instrument of choice. Example, 400 µL water.
- Parameter Value[Derivatization]
- If the sample has been subjected to chemical modification prior to injection, describe the modification. Example, sylilation.
- Parameter Value[Chromatography instrument]
- Add the full name of the instrument used for the Chromatographic part of this assay, including the manufacturer and model number as reported in manufacturer’s brochures, user manuals, or on their website. Example, Shimadzu Nexera UHPLC system.
- Parameter Value[Autosampler model]
- Manufacturer and model number of the autosampler used.
- Parameter Value[Column model]
- Manufacturer, model number and dimensions of the column used. Example, HSS T3 C18 (1.8 μm, 1.0 x 100 mm; Waters).
- Parameter Value[Column type]
- Type or phase of column used. Example, reverse phase.
- Parameter Value[Guard column]
- Type of guard column used.
- Parameter Value[Column model 1]
- Model of first GCGC column.
- Parameter Value[Column type 1]
- Type of first GCGC column.
- Parameter Value[Column model 2]
- Model of second GCGC column.
- Parameter Value[Column type 2]
- Type of second GCGC column.
- Parameter Value[Detector]
- Type, name and manufacturer of detector used.
- Parameter Value[Signal range]
- Range of wavelength of detector used. Example, 200 to 650 nm
- Parameter Value[Resolution]
- Resolution of ditecter used. Example, 2 nm
- Parameter Value[Temperature]
- Temperature of column used.
- Parameter Value[Scan polarity]
- An acquisition mode to which specifies weather polarity is negative, positive or alternating.
- Parameter Value[Scan m/z range]
- The m/z range used in the assay. Example, 100-1000.
- Parameter Value[Instrument]
- Add the full name of the mass spectrometer/detector you used for this LC-MS assay, including the instrument manufacturer and model number as reported in manufacturer’s brochures, user manuals, or on their website. Example, Bruker micrOTOF-Q II.
- Parameter Value[Ion source]
- The ion source where applicable to the instrument, e.g. ESI.
- Parameter Value[Mass analyzer]
- The analyser/detector of the mass fragments generated during the assay. Example, Triple quadrupole.
- Parameter Value[CE instrument]
- The name of the capillary electrophoresis instrument, manufacturer, model.
- Parameter Value[Autosampler model]
- Manufacturer and model number of the autosampler used for the capillary electrophoresis.
- Parameter Value[Column model]
- Manufacturer and model number of capillary column used.
- Parameter Value[Column type]
- Type of capillary column used.
- Parameter Value[DI instrument]
- The name of the direct infusion instrument.
- Parameter Value[FIA instrument]
- The name of the flow injection analysis instrument.
- Parameter Value[Instrument manufacturer]
- The manufacturer of the mass spectrometry imaging instrument.
- Parameter Value[Solvent]
- Name of spray solvent used.
- Parameter Value[Target material]
- Target material used.
- Parameter Value[Spatial resolution]
- Spatial resolution. Example, 10 micrometer
- Parameter Value[Pixel size x]
- Pixel size of X-axis.
- Parameter Value[Pixel size y]
- Pixel size of Y-axis.
- Parameter Value[Max count of pixel x]
- Maximum count of pixel of X-axis.
- Parameter Value[Max count of pixel y]
- Maximum count of pixel of Y-axis.
- Parameter Value[Max dimension x]
- Maximum lenght of aquired image of X-axis.
- Parameter Value[Max dimension y]
- Maximum lenght of aquired image of Y-axis.
- Parameter Value[Inlet type]
- Type of MS inlet used.
- Parameter Value[Detector]
- Type, name and manufacturer of detector used.
- Parameter Value[Detector mode]
- Mode of detector used.
- Parameter Value[Resolving power]
- Resolving power of detector used.
- Parameter Value[Resolving power m/z]
- Resolving power of m/z.
- Parameter Value[Native spectrum identifier format]
- Format of native spectrum identifier.
- Parameter Value[Data file content]
- Content of data file.
- Parameter Value[Spectrum representation]
- Representation type of spectrum.
- Parameter Value[Raw data file format]
- Format of raw data file.
- Parameter Value[Instrument software]
- Name of instrument software used.
- Parameter Value[Instrument software version]
- Version of instrument software used.
- Parameter Value[Line scan direction]
- Direction of first line scan.
- Parameter Value[Line scan sequence]
- Sequence of line scan.
- Parameter Value[Scan pattern]
- Pattern of scan.
- Parameter Value[Scan type]
- Type of scan.
- Parameter Value[Number of scans]
- Number of scan.
- Parameter Value[Sample mounting]
- Mounting method of sample used.
- Parameter Value[Sample preservation]
- Preservation method of sample used. Example, Freezing
- Parameter Value[Tissue modification]
- Modification method of tissue used.
- Parameter Value[Sectioning instrument]
- Name and manufacturer of sectioning instrument used.
- Parameter Value[Section thickness]
- Thickness of section used.
- Parameter Value[Matrix]
- Name and/or type of matrix compound used.
- Parameter Value[Matrix application]
- Application method of matrix compound used.
- Parameter Value[Stain]
- Method of staining.
- Parameter Value[Data processing software]
- Name of data processing software used.
- Parameter Value[Data processing software version]
- Version of data processing software used.
- Parameter Value[Extraction method]
- How a sample was extracted from its source material, e.g. Methanol.
- Parameter Value[NMR tube type]
- Size and type of tube. Example, standard 5 mm glass NMR tube (Wilmad, LabGlass, USA).
- Parameter Value[Solvent]
- Solvent used in the NMR sample, e.g. D2O.
- Parameter Value[Sample pH]
- Sample pH value, e.g. 7.
- Parameter Value[Temperature]
- Sample temperature value with relevant temperature unit.
- Parameter Value[Instrument]
- Add the full name of the instrument you used for the NMR study in this assay, including the model number and its operating frequency. Example, Varian Unity Inova 500 MHz spectrometer.
- Parameter Value[NMR probe]
- Add a full description including the name and type of probe used. This information can be found in the ‘Acquisition Parameter Data File’, ‘acqus.txt’ found within the Bruker raw data file structure, in the field marked ‘$PROBHD=’ Example, 5 mm CPTCI 1H-13C/15N/D Z-GRD.
- Parameter Value[Number of transients]
- The number of scans acquired. This information can be found in the ‘Acquisition Parameter Data File’, ‘acqus.txt’ found within the Bruker raw data file structure, in the field marked ‘$NS=’. Example, 128.
- Parameter Value[Pulse sequence name]
- The pulse sequence program used with a short description. This information can be found in the ‘Acquisition Parameter Data File’, ‘acqus.txt’ found within the Bruker raw data file structure, in the field marked ‘$PULPROG=’ and in the file ‘pulseprogram.txt’. Example, 1D 1H with presaturation (presat).
- Parameter Value[Magnetic field strength]
- Magnetic field strength in Tesla (T), e.g. 11.7.