Genomic Expression Archive
Metadata
MAGE-TAB
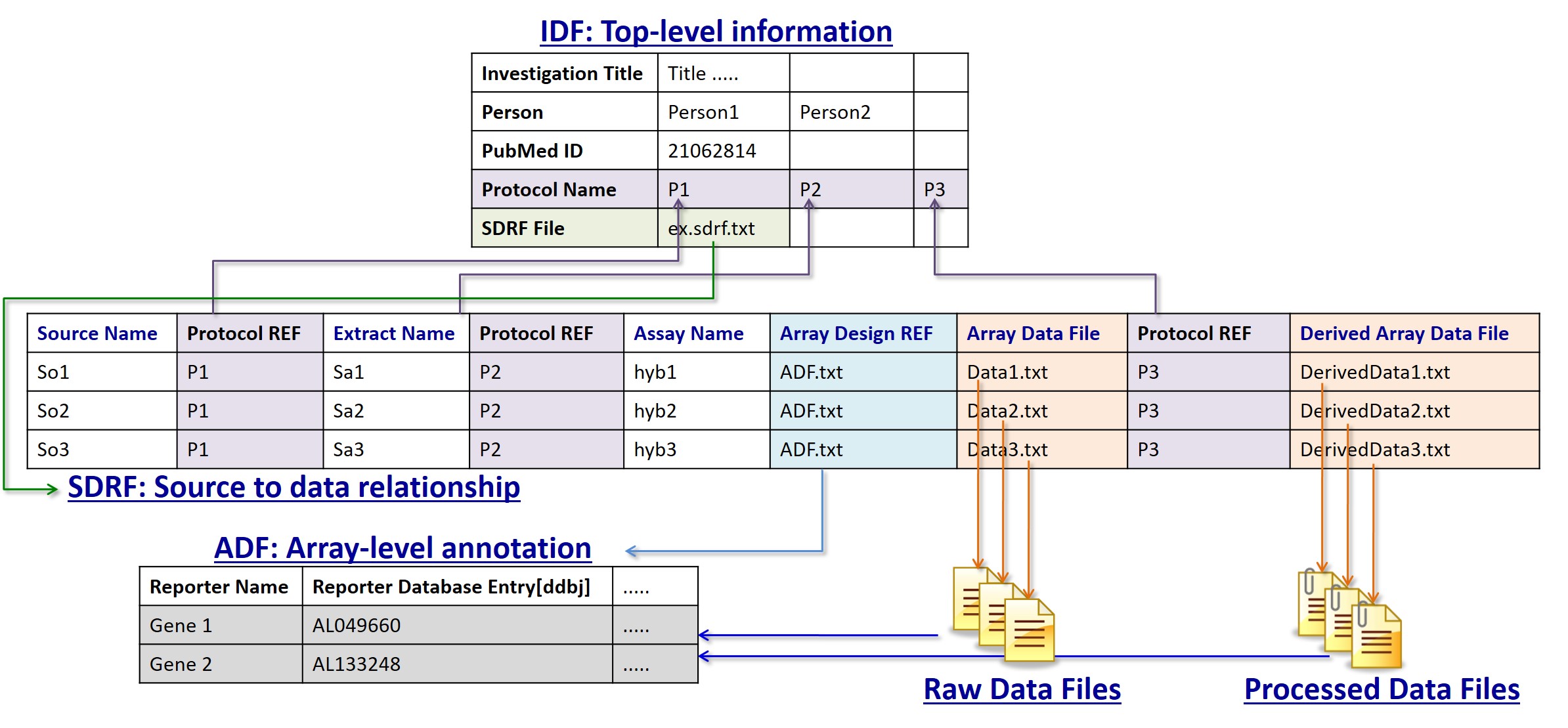
The MicroArray Gene Expression Tabular (MAGE-TAB) format has been developed and adopted by the functional genomics community as a means of representing and communicating data about a functional genomics experiment in a structured and standardised way. The full specification outlines the format.
IDF
The IDF (Investigation Description Format) file is used to give an overview of the experiment, including the experimental design, protocols, publication information and submitter details. IDF examples
IDF fields
The IDF component of a MAGE-TAB document consists of a set of unique tags attached to their corresponding values in a simple tab-delimited text format. For example, “Experiment Description” should be followed by a free-text description of the experiment. Most of the following fields can be used with more than one value, so that multiple protocols, persons, experimental factors etc. can be defined in a single IDF file. In these cases, the different “objects” are separated by tabs. Those fields which can contain only one value are indicated below.
- Investigation Title
- The overall title of the investigation. This tag can only have one value.
- Experimental Design
- The experiment design types which are applicable to this study. These terms should come from controlled terms.
- Experimental Factor Name
- A user-defined name for each experimental factor studied by the experiment. These experimental factors represent the variables within the investigation (e.g. growth condition, genotype, organism part). The actual values of these variables will be listed in the SDRF file, in “Factor Value[
]" columns. GEA copies the Factor Value names in SDRF to IDF. - Experimental Factor Type
- A term describing the type of each experimental factor. GEA uses the Experimental Factor Name for the Experimental Factor Type.
- Person Last Name
- The last name of each person associated with the experiment.
- Person First Name
- The first name of each person associated with the experiment.
- Person Mid Initials
- The middle name of each person associated with the experiment.
- Person Affiliation
- The organization affiliation for each person associated with the experiment.
- Person Roles
- The role(s) performed by each person. GEA only permits “submitter” role.
- Public Release Date
- The date on which the experimental data was released. This tag can only have one value.
- PubMed ID
- The PubMed IDs of the publication(s) associated with this investigation (where available).
- Publication DOI
- A Digital Object Identifier (DOI) for each publication (where available). When PubMed ID and DOI are available, use PubMed ID.
- Experiment Description
- A short paragraph describing the experiment as free-text. This tag can only have one value. The text should clearly explain what you did in your experiment - this will help the curation team to check and process your MAGE-TAB document.
- Protocol Name
- The names of the protocols used within the MAGE-TAB document. These will be referenced in the SDRF in the “Protocol REF” columns by procotol’s temporary IDs (e.g. ESUB000350_Protocol_1) or accessions (e.g. P-GEAD-100). GEA does not support protocol reference across experiments.
- Protocol Type
- The type of the protocol. Pre-defined types are as follows. Necessary protocol sets are different between Microarray and Sequencing experiments.
- array scanning and feature extraction protocol
- conversion protocol
- dissection protocol
- growth protocol
- high throughput sequence alignment protocol
- normalization data transformation protocol
- nucleic acid extraction protocol
- nucleic acid hybridization to array protocol
- nucleic acid labeling protocol
- nucleic acid library construction protocol
- nucleic acid sequencing protocol
- sample collection protocol
- treatment protocol
- Protocol Description
- A free-text description of the protocol. This text is included in a single tab-delimited field.
- SDRF File
- The name(s) of the SDRF file(s) accompanying this IDF file. GEA automatically embeds the value.
- Comment[<user-defined tag>]
- A user- or archive-defined value which is associated with the investigation. Following Comment tags are used by GEA.
- Comment[GEAAccession]
- Comment[AEExperimentType]
- Comment[Number of channel]
- Comment[Array Design REF]
- Comment[BioProject]
- Comment[Public Release Date]
- Comment[Last Update Date]
- Comment[AdditionalFile:TXT]
SDRF
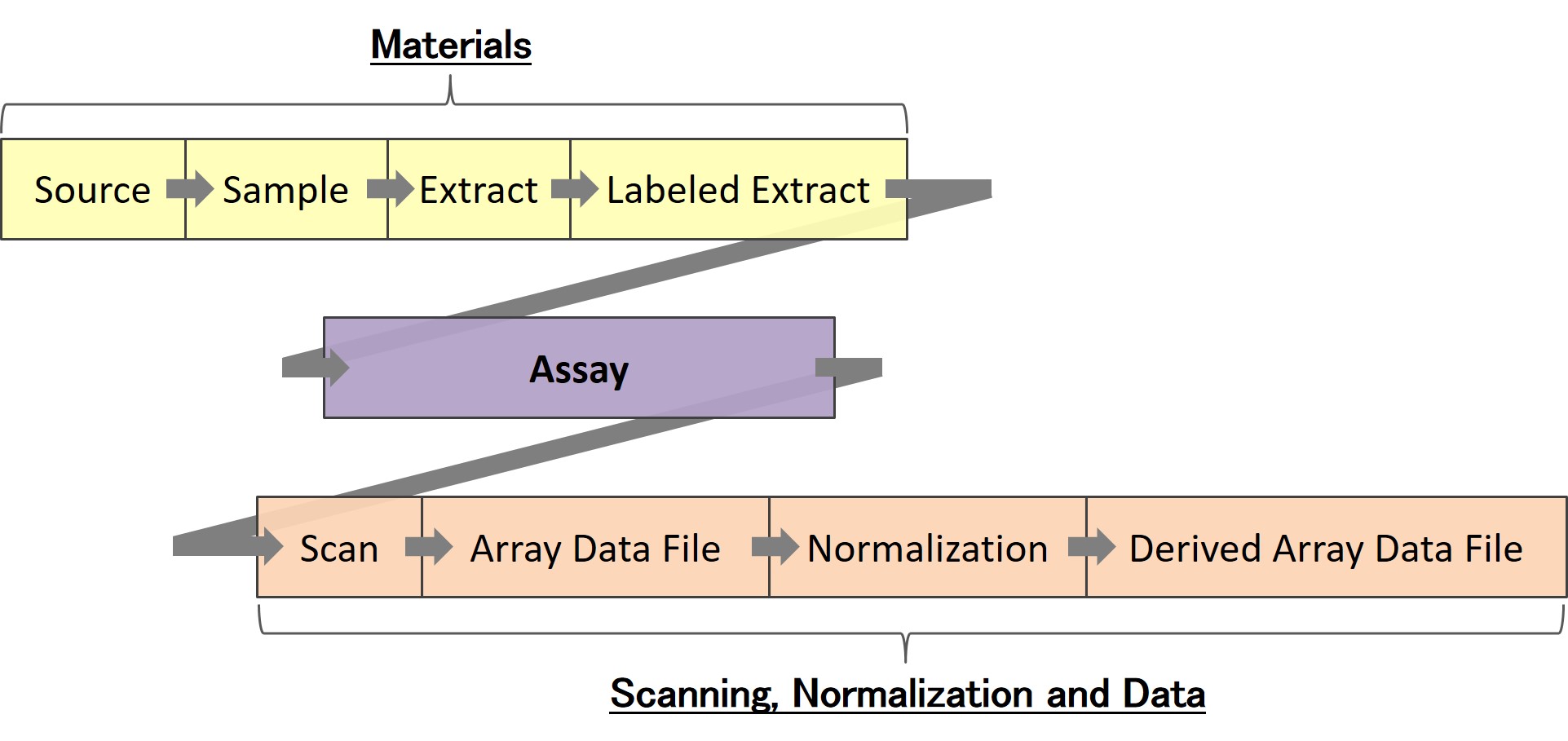
The SDRF (Sample and Data Relationship Format) describes the sample characteristics and the relationship between samples, arrays, data files etc. The information in the SDRF is organized so that it follows the natural flow of a functional genomics experiment. It begins with describing your samples and finishes with the names of the data files generated from the analysis of the experiment results. For single-channel microarray data one row in the SDRF is equal to one hybridization. For two-channel microarray data one row is equal to one channel. For sequencing submissions you generally create one row per raw file (Run) including paired sequencing. SDRF examples
SDRF sections
- Protocols
- In most cases, each treatment within an experiment will be represented simply by a Protocol REF column containing references to the Protocol Names defined in the IDF.
In un-registered submission, protocols in IDF are assigned temporal IDs (e.g., ESUB000001_Protocol_1) and are referenced by SDRF by these IDs.
| Protocol Name | ESUB000500_Protocol_1 | ESUB000500_Protocol_2 |
| Protocol Type | nucleic acid extraction protocol | labelling protocol |
The SDRF describing the use of these protocols might look as follows:
| Protocol REF | Protocol REF |
| OSUB000500_Protocol_2 | OSUB000500_Protocol_3 |
- Sources
- Sources are the starting material for the experiment. The section starts with a Source Name column, which will typically be followed by several Characteristics columns.
In the GEA submission system, a sample_name of relevant BioSample is commonly used for Names of Source, Extract, Labeled Extract and Assay columns.
| Source Name | Characteristics[organism] | Characteristics[ecotype] |
| Arabidopsis control for heat stress_rep 1 | Arabidopsis thaliana | Col-0 |
Additional columns which may be used to annotate Sources are: Provider and Comment.
- Extracts
- Extracts refer to the extracted nucleic acid used in the experiment.
| Source Name | Protocol REF | Extract Name | Material Type |
| Arabidopsis control for heat stress_rep 1 | OSUB000500_Protocol_2 | Arabidopsis control for heat stress_rep 1 | total RNA |
Columns which may be used to annotate Extracts are: Characteristics[], Material Type and Comment.
- Labeled Extracts
- The Labeled Extracts in an experiment are those materials which have been conjugated to a label, prior to hybridization on an array. Typically there is only one labeled extract step. For microarray experiment submission to GEA, a Label column must be included with the Labeled Extract Name column to indicate which label (and therefore scanner channel) corresponds to which sample:
| Extract Name | Material Type | Protocol REF | Labeled Extract Name | Label |
| Arabidopsis control for heat stress_rep 1 | total RNA | OSUB000500_Protocol_3 | Arabidopsis control for heat stress_rep 1 | biotin |
Columns which may be used to annotate Labeled Extracts are: Characteristics[], Material Type and Comment.
- Assays
- The hybridization of labeled extract to an array or the sequencing of a library is a key step in the SDRF, since it connects the “materials” section of the SDRF to the “data” section. For an array submission to GEA, an Array Design REF column must be included with the Assay Name column, indicating which array design was used
| Labeled Extract Name | Label | Assay Name | Technology Type | Array Design REF |
| Arabidopsis control for heat stress_rep 1 | biotin | Arabidopsis control for heat stress_rep 1 | array assay | A-AFFY-2 |
It is also possible to use Comment columns to annotate both “Assay Name” columns. Note that the values in Assay Name columns may be used in Data Matrix files to link columns of data to individual assays.
- Raw Data Files
- The raw data files generated by an investigation should be listed in an Array Data File column following the Assay Name column.
| Assay Name | Technology Type | Array Design REF | Array Data File |
| Arabidopsis control for heat stress_rep 1 | array assay | A-AFFY-2 | Arabidopsis_control_rep_1.CEL |
Comment columns can be used to add information relating to the raw files.
- Processed Data Files
- The processed data files which have been derived from the raw data should be listed in a Derived Array Data File column for both microarray- and sequencing-based experiments. Note that this generally only applies to processed data arranged into one file per assay. If your files contain processed data columns for more than one assay, you should reformat these into the Data Matrix format and include them instead in a Derived Array Data Matrix File column.
| Assay Name | Technology Type | Array Design REF | Array Data File | Derived Array Data File |
| Arabidopsis control for heat stress_rep 1 | array assay | A-AFFY-2 | Arabidopsis_control_rep_1.CEL | Arabidopsis_control_rep_1.processed.txt |
- Factor Values
- The factor values for an experiment are the values of the variables under investigation. For example, an experiment studying the effect of different temparature (heat stress) on a cell culture would have “temparature” as an experimental variable with “Unit” column to indicate the unit.
| Source Name | Comment[description] | Assay Name | Factor Value[temparature] | Unit[temperature unit] |
| Arabidopsis control for heat stress rep 1 | Arabidopsis control for heat stress, biological replicate 1 | Arabidopsis control for heat stress rep 1 | 22 | degree_C |
| Arabidopsis heat stress rep 1 | Arabidopsis heat stress, biological replicate 1 | Arabidopsis heat stress rep 1 | 37 | degree_C |
Note that there is inevitably duplication between factor values and values entered elsewhere in the SDRF. It is particularly common to have the “Factor Value” column duplicate either a Characteristics[] column.
Factor Value columns must be placed at the end (i.e., the far right) of the SDRF.
GEA lists the variable in the IDF as “Experimental Factor Name” with associated Type (usually same with the name).
| Experimental Factor Name | temperature |
| Experimental Factor Type | temperature |
SDRF columns
- Source Name
- Used as an identifier within the MAGE-TAB document. This column contains user-defined names (by default, a sample name of corresponding BioSample is used) for the Source materials. The following columns can be used to annotate “Source Name” columns:
- Sample Name
- Used as an identifier within the MAGE-TAB document. This column contains user-defined names (by default, a sample name of corresponding BioSample is used) for each Sample material. The following columns can be used to annotate “Sample Name” columns:
- Extract Name
- Used as an identifier within the MAGE-TAB document. This column contains user-defined names (by default, a sample name of corresponding BioSample is used) for each Extract material. The following columns can be used to annotate “Extract Name” columns:
- Labeled Extract Name
- Used as an identifier within the MAGE-TAB document. This column contains user-defined names (by default, a sample name of corresponding BioSample is used) for each Labeled Extract material. The following columns can be used to annotate “Labeled Extract” Name columns:
- Assay Name
- Used as an identifier within the MAGE-TAB document. This column contains user-defined names (by default, a sample name of corresponding BioSample is used) for each Assay. The following columns can be used to annotate “Assay Name” columns:
- Array Design REF (required for array submissions)
- Technology Type (“array assay” for microarray and “sequencing assay” for sequencing experiment)
- Comment[]
All “Assay Name” columns must be followed by a “Technology Type” column.
- Scan Name
- Used as an identifier within the MAGE-TAB document. This optional column contains user-defined names for each Scan event. The following columns can be used to annotate “Scan Name” columns:
- Normalization Name
- Used as an identifier within the MAGE-TAB document. This optional column contains user-defined names for each Normalization event. The following columns can be used to annotate “Normalization Name” columns:
- Array Data File
- This column contains a list of raw data files, one for each row of the SDRF file, linking these data files to their respective assays. The following columns can be used to annotate “Array Data File” columns:
- Derived Array Data File
- This column contains a list of microarray and sequencing processed data files, one for each row of the SDRF file, linking these data files to their respective assays. The following columns can be used to annotate “Derived Array Data File” columns:
- Array Data Matrix File
- This column contains a list of microarray and sequencing raw data matrix files, where data from multiple assays is stored in a single file, and the data mapped to each assay via the Data Matrix format itself. The following columns can be used to annotate “Array Data Matrix File” columns:
- Derived Array Data Matrix File
- This column contains a list of microarray and sequencing processed data matrix files, where data from multiple assays is stored in a single file, and the data mapped to each assay (or scan, or normalization) via the Data Matrix format itself. The following columns can be used to annotate “Derived Array Data Matrix File” columns:
- Array Design REF
- This column contains references to the array design used for each assay. For GEA submissions this should be an ArrayExpress/GEA accession number, e.g. “A-AFFY-2”. An array design accession can be found at ArrayExpress array design search page. The following columns can be used to annotate “Array Design REF” columns:
- Protocol REF
- This column contains references to Protocol Names defined in the IDF. Before registration, a protocol is referenced by its temporal ID like “ESUB000500_Protocol_1”. After registration, this ID is replaced by a protocol accession number like “P-GEAD-100”. The following columns can be used to annotate “Protocol REF” columns:
- Characteristics[<category term>]
- Controlled vocabulary term or measurement. Used as an attribute column following Source Name, Sample Name, Extract Name, or Labeled Extract Name. This column contains terms describing each material according to the characteristics category indicated in the column header. For example, a column headed “Characteristics[organism]” would contain organism names, for example “Homo sapiens”. These terms may be user-defined (the default), from an external ontology source (NCBI Taxonomy for organism name), or a measurement (indicated using a Unit[] column).
- Provider
- Used as an attribute column following Source Name. A free-text string identifying the organization or person from which the Source was obtained.
- Material Type
- Controlled vocabulary term. Used as an attribute column following Source Name, Sample Name, Extract Name, or Labeled Extract Name. This column contains the following controlled terms describing the type of each material.
- total RNA
- polyA RNA
- cytoplasmic RNA
- nuclear RNA
- genomic DNA
- protein
- other
- Label
- Controlled vocabulary term. Used as an attribute column following Labeled Extract Name. The label compound which is conjugated to an Extract to create the Labeled Extract. Examples: Cy3, Cy5, biotin, alexa_546. The following columns can be used to annotate Label columns:
- Technology Type
- Controlled vocabulary term. Used as an attribute column following Assay Name. GEA auto-fills “array assay” for array submission and “sequencing assay” for sequencing submission. The following columns can be used to annotate Technology Type columns:
- Factor Value[<experiment factor name>]
- Controlled vocabulary term or measurement. This column contains terms describing the experimental factor values (i.e., variables) for each row of the SDRF. The Experimental Factor Name to which it pertains should be indicated in the column heading. In a SDRF template file provided in the submission system, enter a variable name by overwriting “enter experiment factor name here” of “Factor Value[enter experiment factor name here]”.
Example SDRF:
| Factor Value[tissue] |
| gall bladder |
| kidney |
| liver |
| intestine |
| pancreas |
In the example above, the column terms would be treated as describing tissues. For more precise control over the treatment of these terms, the optional form “Factor Value” is available, e.g. “Factor Value[growthconditionEF](Nutrients)”.
When a combination of more than one variables are studied, factor values are described by more than one columns. Example SDRF:
| Factor Value[compound] | Factor Value[dose] | Unit[molar mass unit] |
| none | ||
| potassium cyanide | 25 | micromolar |
| potassium cyanide | 35 | micromolar |
| potassium cyanide | 50 | micromolar |
The factor name shoud be defined in accompanying IDF (GEA auto-fills the factor name in SDRF to IDF):
| Experimental Factor Name | tissue |
| Experimental Factor Type | tissue |
- Performer
- Used as an attribute column following Protocol REF. The name of the researcher or center where the protocol was carried out.
- Date
- Used as an attribute column following Protocol REF. The date (and time, where available) upon which the protocol was performed, in the following format: YYYY-MM-DD.
- Parameter Value[<protocol parameter>]
- Used as an attribute column following Protocol REF columns. This column contains values for the protocol parameters referenced in the column header. The following columns can be used to annotate Parameter Value[] columns:
For example, if a Protocol Name “Array Hybridization” is defined in the accompanying IDF, with Protocol Parameters “hyb temp”, the following would be valid:
| Protocol REF | Parameter Value[hyb temp] | Unit[temperature unit] |
| Array Hybridization | 55 | degree celsius |
- Unit[<unit category>]
- Controlled vocabulary term. Used as an attribute column following Characteristics[], Factor Value[] or Parameter Value[]. This column contains terms describing the unit(s) to be applied to the values in the preceding column. The type of unit is included in the column heading, e.g. “Unit[volume unit]”. These unit types should correspond to Unit subclasses from EFO. The following columns can be used to annotate Unit[] columns:
The Term Source REF column in this case would point to the ontology from which the Unit terms are taken.
- Description
- Used as an attribute column following Source Name, Sample Name, Extract Name, or Labeled Extract Name. A free-text description to be attached to the corresponding material. To be used sparingly, if at all - most annotations should be provided using controlled vocabulary terms, using Characteristics[] columns.
- Term Source REF
- Used as an attribute column following any controlled vocabulary column (e.g., Characteristics[]), or column allowing reference of external entities (e.g., Protocol REF). This column contains references to ontology or database Term Sources defined in the IDF, and from which the values in the previous column were taken. The following columns can be used to annotate Term Source REF columns.
- Term Accession Number
- Used as an attribute column following Term Source REF columns. This column contains the accession numbers from the term source used to identify the ontology or database terms in question. For example:
| Source Name | Characteristics[disease] | Term Source REF | Term Accession Number |
| Sample 1 | acute lymphoblastic leukemia | EFO | EFO_0000220 |
(This example relies on the EFO Term Source having been pre-defined in the IDF accompanying the SDRF.) At the point of submission, GEA does not require ontology usage as ArrayExpress.
- Comment[<comment name>]
- This column can be used to annotate the main graph node and edge columns listed above. It is included as an extensibility mechanism, and should not generally be used to encode meaningful biological annotation. The column heading should contain a name for the type ofvalues included in the column.