Genomic Expression Archive
MAGE-TAB example
IDF Example
Full IDF example in spreadsheet (Microarray) Full IDF example in spreadsheet (NGS)
| Comment[GEAAccession] | E-GEAD-369 | |
| MAGE-TAB Version | 1.1 | |
| Investigation Title | Transcriptional profiling of Arabidopsis leaves exposed to elevated temperature | |
| Experiment Description | We investigated genome-wide … biological replicates were analyzed. | |
| Experimental Design | stimulus or stress design | |
| Experimental Factor Name | temperature | |
| Experimental Factor Type | temperature | |
| Person Last Name | Test | |
| Person First Name | Tarou | |
| Person Affiliation | DDBJ Center, National Institute of Genetics | |
| Person Roles | submitter | |
| Public Release Date | 2018-06-22 | |
| PubMed ID | 29040613 | |
| Protocol Name | P-GEAD-535 | … |
| Protocol Type | sample collection protocol | … |
| Protocol Description | Plants were sown on GM medium … 22 degrees celsius. | … |
| SDRF File | E-GEAD-369.sdrf.txt | |
| Comment[Number of channel] | single-channel | |
| Comment[Array Design REF] | A-AFFY-2 | |
| Comment[AEExperimentType] | transcription profiling by array | |
| Comment[BioProject] | PRJDB3647 | |
| Comment[Public Release Date] | 2018-06-22 | |
| Comment[Last Update Date] | 2018-06-22 |
A full listing of all supported IDF tags can be found in the IDF section.
SDRF Conceptual Examples
Full SDRF example in spreadsheet (Microarray) Full SDRF example in spreadsheet (NGS)
- Example 1: Raw and processed microarray data files for each sample
- Example 2: Two-color microarray
- Example 3: Next-generation sequencing raw DRA and processed data for each sample
In the below examples, the simplified SDRF (only conceptually-important columns are represented) has been represented by a row in the corresponding table. Column headings in blue denote graph node identifier columns (names).
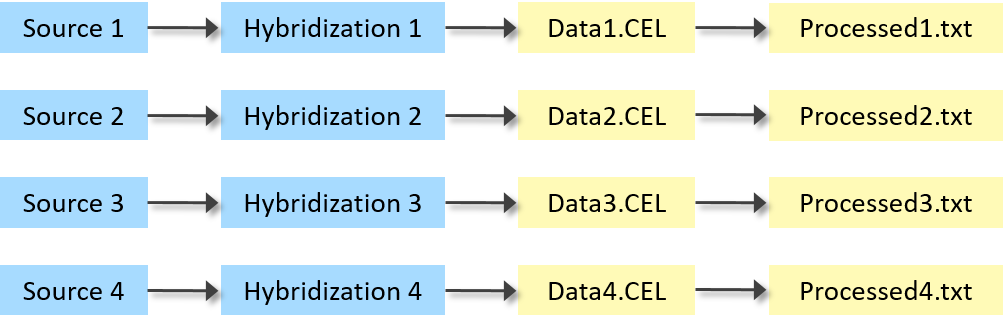
Example 1: Raw and processed microarray data files for each sample
A simple microarray experiment. Each sample has one raw data file and one processed data file.
| Source Name | Protocol REF | Assay Name | Array Data File | Derived Array Data File |
| Source 1 | P-GEAD-10 | Hybridization 1 | Data1.CEL | Processed1.txt |
| Source 2 | P-GEAD-10 | Hybridization 2 | Data2.CEL | Processed2.txt |
| Source 3 | P-GEAD-10 | Hybridization 3 | Data3.CEL | Processed3.txt |
| Source 4 | P-GEAD-10 | Hybridization 4 | Data4.CEL | Processed4.txt |
Raw and processed microarray data files for each sample
Example 2: Two-color microarray
| Source Name | Extract Name | Labeled Extract Name | Label | Assay Name |
| Source 1a | Extract 1a | LabeledExtract 1a Cy3 | Cy3 | Hybridization 1 |
| Source 1b | Extract 1b | LabeledExtract 1b Cy5 | Cy5 | Hybridization 1 |
| Source 2a | Extract 2a | LabeledExtract 2a Cy3 | Cy3 | Hybridization 2 |
| Source 2b | Extract 2b | LabeledExtract 2b Cy5 | Cy5 | Hybridization 2 |
| Source 3a | Extract 3a | LabeledExtract 3a Cy3 | Cy3 | Hybridization 3 |
| Source 3b | Extract 3b | LabeledExtract 3b Cy5 | Cy5 | Hybridization 3 |
Two-color microarray
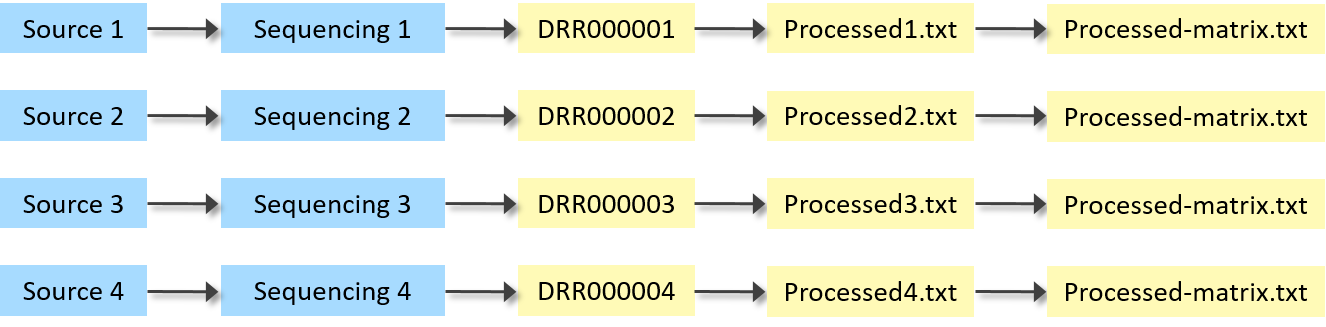
Example 3: Raw and processed sequencing data file for each sample, and a processed matrix file
A simple next-generation sequencing experiment. One raw DRA data file and one processed data file for each sample, and one processed matrix file for all samples.
| Source Name | Protocol REF | Assay Name | Array Data File | Derived Array Data File | Derived Array Data Matrix File |
| Source 1 | P-GEAD-20 | Sequencing 1 | DRR000001 | Processed1.txt | Processed-matrix.txt |
| Source 2 | P-GEAD-20 | Sequencing 2 | DRR000002 | Processed2.txt | Processed-matrix.txt |
| Source 3 | P-GEAD-20 | Sequencing 3 | DRR000003 | Processed3.txt | Processed-matrix.txt |
| Source 4 | P-GEAD-20 | Sequencing 4 | DRR000004 | Processed4.txt | Processed-matrix.txt |
Raw and processed sequencing data file for each sample, and a processed matrix file
MAGE-TAB Real-world Examples
Full SDRF example in spreadsheet.
Following table lists the real-world MAGE-TAB example files of the microarray and sequencing data deposited into the ArrayExpress.
| Accession Number | IDF Text | SDRF Text | Experiment Summary | Platform |
| E-TABM-18 | idf text | sdrf text | Transcription profiling of 35 different Arabidopsis thaliana ecotypes | Affymetrix |
| E-TABM-985 | idf text | sdrf text | Transcription profiling by array of mouse testis development, dye swap design | Compugen |
| E-MEXP-2665 | idf text | sdrf text | ChIP-chip by array of mouse cell lines and fresh tissues | Multiple platforms |
| E-TABM-855 | idf text | sdrf text | Genotyping of human lymphoblastoid cell lines from pairs of HapMap indiviudals | Illumina GoldenGate SNP array |
| E-MTAB-371 | idf text | sdrf text | ChIP-seq of human myofibroblasts | Illumina Sequencing |
| E-MTAB-6311 | idf text | sdrf text | Transcription changes in zebrafish embryos of different developmental ages after exposure to cyanide (two factor values) | Affymetrix |
| E-GEOD-26444 | idf text | sdrf text | Deep sequencing-based analysis of the anaerobic stimulon | SOLiD sequencing |
| E-GEOD-24366 | idf text | sdrf text | Global changes following N-deprivation in Chlamydomonas | 454 sequencing |
MAGE-TAB real-world examples