Genomic Expression Archive
Array design
Array design format (ADF) file
The ADF (Array Design Format) file captures information about a microarray chip. The file contains two section, separated by a [main] tag:
- A multi-row metadata header (e.g. who submitted the array design, how the probes or composite “probe sets” were designed); followed by
- A table with columns of annotation for each probe/probe set in rows, e.g. unique probe identifier, probe sequences, probe coordinate location (on spotted arrays), cross-references to other databases, where the probes map to in the genome, and so on.
The ADF file is always in tab-delimited text (*.txt) format, and will open in any spreadsheet program for viewing or editing.
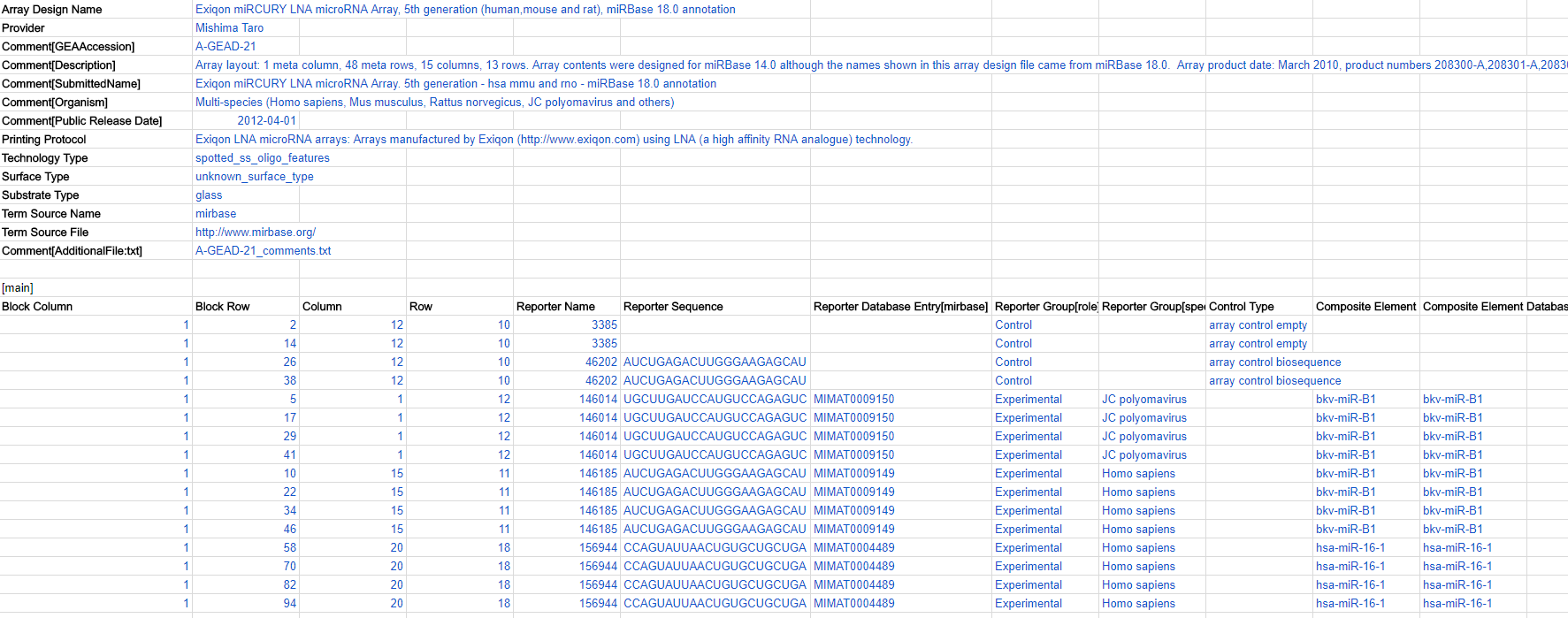
Here is a snippet of an ADF document:
ADF metadata header
Each row (field) of metadata starts with a heading (bold in the table below), each appearing only once and coming from a controlled vocabulary. The headings would be pre-filled for you when you download one of our ADF templates so you don’t need to insert them from scratch. Please do not edit the headings, but fill in each field as much as possible, like the blue text below:
A glossary of the ADF header fields (* denotes mandatory fields):
| No. | ADF header field | What is it? | Allowed values | Example |
|---|---|---|---|---|
| 1 | * Array Design Name | An informative title of the array design. Should include the name of the manufacturer (e.g. Agilent) or the lab which designed it, the species, version number, purpose of the array (e.g. genotyping), the number of probes/features (e.g 450k) | Agilent human micoRNA microarray miRBase Release 14.0, 8x15k (GridName 029297, 82 cols x 192 rows) | |
| 2 | Version | Version (or revision) number of the array design. | version 3.0 | |
| 3 | * Provider | Name of the array design’s submitter. | Mishima Taro | |
| 4 | * Comment[Organism] | The species from which the probe sequences were derived from. | Must use scientific name in NCBI Taxonomy. Separate names by semicolon for multi-species arrays. | Homo sapiens |
| 5 | * Comment[Description] | A concise description of how the microarray is designed and what it is for. Include URLs to stable online resources if possible. | This custom commercial microarray consists of 84881 (86205 total including 1324 control probes) 60-mer oligonucleotide probes derived from the genomes and EST database of Emiliania huxleyi strain (… etc…) | |
| 6 | * Comment[Public Release Date] | Enter “Hold” to keep pre-published array designs private (inform us to release). Otherwise, insert the date of submission. | Must use YYYY-MM-DD format for submission date. | 2014-04-25 |
| 7 | Printing Protocol | How the probes were printed on the array. Include URLs to stable online resources if possible. Especially important if you are submitting a custom array printed with non-propreitary methods. | The 60-mer oligonucleotides were synthessized in situ using Agilent inkjet SurePrint technology. Four arrays were printed on each 1 x 3-inch glass slide (…etc…) | |
| 8 | Technology Type | Printing/synthesis technology used when creating the microarray. | “in situ oligo features”, “spotted antibody features”, “spotted colony features”, “spotted ds DNA features”, “spotted protein features”, “spotted_ss_PCR_amplicon_features”, “spotted_ss_oligo_features” | in situ oligo features |
| 9 | Surface Type | The chemical coating on the surface of the substrate (see below). Probes are immobilised by the coating. | polylysine, aminosilane, “other surface type” | polylysine |
| 10 | Substrate Type | The material/substrate of the microarray. | glass, nitrocellulose, nylon, silicon, “other substrate type” | glass |
| 11 | Sequence Polymer Type | The polymer that makes up the probes. | DNA, RNA, protein | DNA |
| 12 | Term Source Name | In the ADF table, for each cross-reference provided, e.g. Reporter Database Entry [uniprot], the cross-referenced resource’s name needs to be entered here, so the resource can be looked up via the URL in Term Source File (see header number 13 below). | External database names from this allowed list | uniprot |
| 13 | Term Source File | The URL where an external database resource mentioned in Term Source Name (see header number 12 above) is hosted. | A valid URL. | http://www.uniprot.org |
ADF table
Just like the metadata header fields, column headings in the ADF table come from a controlled vocabulary. Please use one of the ADF templates, where correct headings have been prefilled for you, saving you inserting them from scratch. Please don’t change the column headings in the template.
- composite_element_array_ADF_template.xls
- commercial_array_header_template.xls
- reporter_only_array_ADF_template.xls
- spotted_array_ADF_template.xls
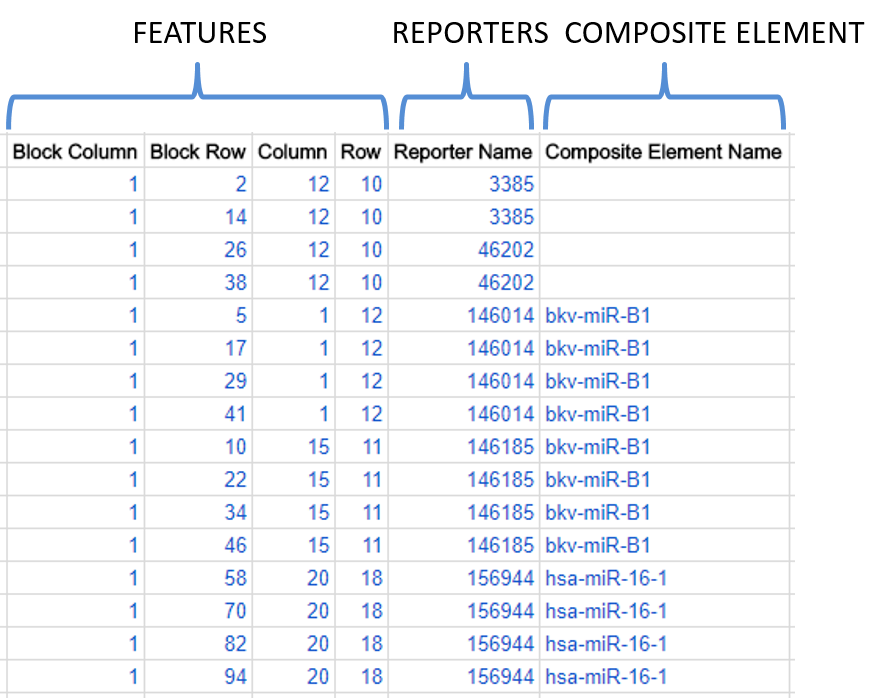
Features, reporters and composite elements
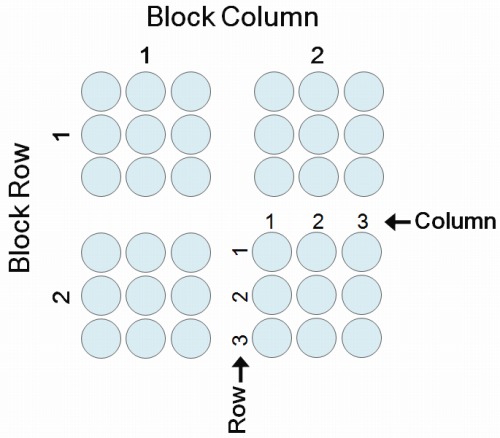
Features (for spotted arrays only):
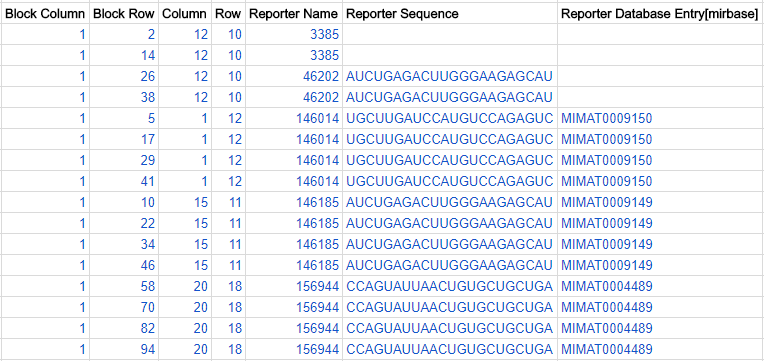
Each spot on the array is called a feature. The position of each feature is described by 4 coordinates, each has its dedicated column in the ADF table: Block Column, Block Row, Column and Row. Each set of 4 coordinates represents a unique position on the microarray, so features cannot be duplicated on an array. Include all features in your ADF file, even if there is nothing spotted there (e.g. control spots). Here is a schematic diagram showing the 4 coordinates:
Reporters (mandatory, for all arrays):
This is what is usually referred to as a “probe”. Each reporter has a particular sequence and a defined role (experimental or control). Give each reporter a unique name in the Reporter Name column, i.e. each name in the column should only appear once, except for spotted arrays (arrays with “features”, see above), where the same probe can be printed at multiple locations on the same array, so the same reporter name can be repeated at different feature locations.
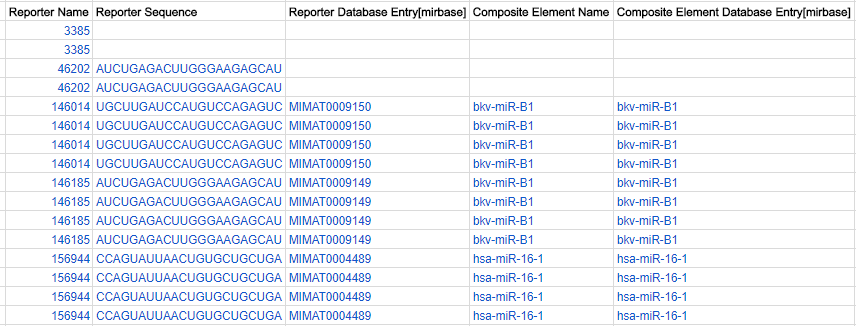
Composite elements (optional, for all arrays):
These allow you to provide an additional level of information showing which reporters are related, e.g. derived from the same gene or transcript. E.g. if you have different reporters that are designed from two exons of a gene, you can indicate that the reporters belong to the same “composite element” (i.e. a “composite” collation of reporters). Give each composite element a unique name in the Composite Element Name column. Affymetrix array designs will always have composite elements because they are based on “probe sets” and not simply “probes”.
Here is a snippet from a hypothetical example of an array design, showing sixteen features, five reporters and two composite elements:
Annotation of reporters
Annotate each reporter by providing its sequence, cross-referenced accessions in external databases (e.g. RefSeq accession numbers of the cDNA sequences from which the reporters were designed), and/or its genomic mapping location. Use the Reporter Sequence column for the sequence, and one or more Reporter Database Entry [xxx] columns for the cross-references and mapping location.
If a reporter appears multiple times in an ADF table, e.g. for spotted arrays where the same reporter is printed at different locations on the microarray, annotation for the same reporter must be consistent (i.e. identical) across the entire ADF table.
For reporters with no cross-references, e.g. a blank control spot, leave annotation blank. Do not put down custom values such as “NA”, “empty” or “unmapped”, as curators will validate the annotation values against expected formats and custom values will not be accepted.
Reporter Sequence:
Use single-letter IUPAC codes for all arrays (including peptide arrays). All experimental reporters and control reporters of type “array control biosequence” should have sequence annotation. In contrast, control reporters which are empty (e.g. nothing spotted on the array), buffer-only or label-only should not have sequence annotation. See section below for more details on experimental and control reporters.
Cross references to external databases:
Look up the abbreviated name of the cross-referenced database from this allowed list, and insert it inside the square brackets of the column heading Reporter Database Entry [xxx], e.g. Reporter Database Entry [genbank]. Then, fill the column with the appropriate accession. You can enter multiple accessions from the same database by separating them with semicolons, e.g. AJ12345;BX45678. For annotations from different database sources, they must be provided in separate Reporter Database Entry [xxx] columns. Don’t forget to add the database name and URL to the Term Source Name and Term Source File fields of the ADF header.
If the database you would like to cite is not already on the list, please contact us with the database URL and we will look into adding it to the list of allowed databases.
We rely on accession numbers and not free-text names, because accessions are usually stable and traceable (even for deprecated ones). Morever, each database usually enforces a certain format for the accessions, which makes computational data-mining a lot easier. E.g. NCBI RefSeq curated cDNAs always have accessions start with NM_ followed by a few digits.
Genomic mapping location:
Instead of an abbreviated database name, enter chromosome_coordinate: inside the square brackets of Reporter Database Entry [xxx], e.g. Reporter Database Entry [chromosome_coordinate:GRCh38]. Try to use official genome assembly names from public databases (ENA, GenBank or DDBJ), e.g GRCh38 for the human genome assembly. Genomic coordinates in the column should follow this format: chrName:start position-end position, e.g. chr1:1234-5678.
This is how the hypothetical reporter annotations would be like. Notice how you can enter multiple accessions from the same database by separating them with semicolons:
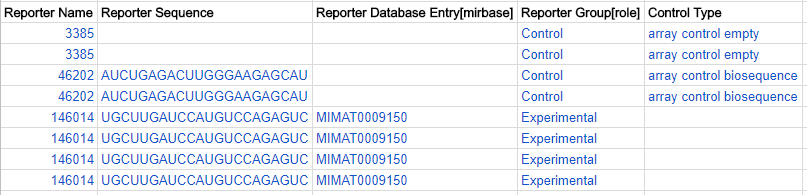
Repoter role - experimental or control
For each reporter, enter its role: experimental or control in the Reporter Group [role] column.
For each control reporter, describe what type of control it is in the Control Type column. (Do not fill in this column for any experimental reporters.) The allowed control type values taken from Experimental Factor Ontology (EFO). Here is a glossary:
| Allowed terms | Meaning |
|---|---|
| array control biosequence | E.g. a spiked sequence from E. coli for a human microarray. Reporter Sequence must be provided. |
| array control buffer | Buffer spotted on the array. Do not provide Reporter Sequence. |
| array control empty | Nothing spotted on the array (blank). Do not provide Reporter Sequence. |
| array control genomic DNA | E.g. salmon sperm DNA |
| array control label | Landing lights. Do not provide Reporter Sequence. |
| array control reporter size | Size standards. |
| array control spike calibration | E.g. the same spike sequence introduced at varying concentrations |
| array control design | (when none of the above applies, i.e. a control was used, but no further details available) |
Here is an example of reporter roles in the ADF table:
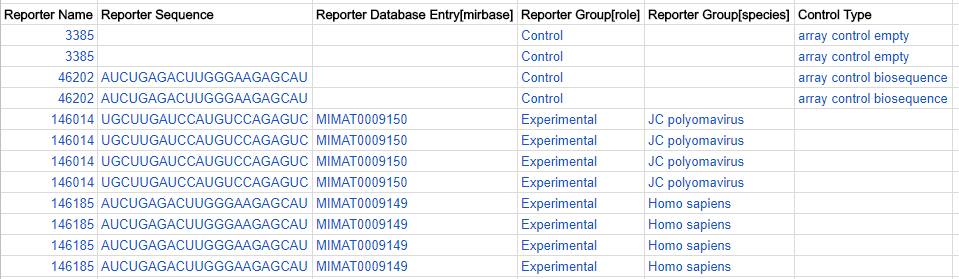
Reporter grouping by species (multi-species ADFs only)
Where reporters were designed from more than one species, you can indicate the species source in the Reporter Group [species] column. Use species name from Latin binomial nomenclature where possible, e.g. Homo sapiens.
An example of a microarray with probes derived from viral and human microRNA sequences:
Annotation of composite elements
Annotation of composite elements is usually in the form of cross-references to external databases, in a very similar way as for reporters, using one or more Composite Element Database Entry [xxx] columns. In addition, a Composite Element Comment column can be added to insert free-text comments or descriptions for each element.