Genomic Expression Archive
Submit microarray data
Pre-submission checklist
Two-color microarray experiment
At the moment GEA submission interface only supports one type of two-color workflow (see example here), where two samples are connected with one common raw data file, which includes both channels. If you select the dual-channel option in the IDF tab, it will expect that you provide one file for the two samples that were hybridized together. Some recent two-color microarray technologies generate two separate raw data files (usually one for each channel), which will cause validation to fail (if you connect a single file per sample). If you have separate files for each channel, please contact GEA team.
Single-cell sequencing experiment
Refer to Single-cell submission guide. Please contact GEA team to upload any additional files for custom spike-ins or to facilitate data analysis.
More than one technology per experiment
GEA will ask you for the technology and name of the array, and applies it to the whole submission. If you have used different types of technologies for the same set of samples, we ask you to create separate submissions. Please make sure that the submissions have distinct titles (even though they may belong to the same study), in order to avoid mistakes. If you have samples from more than one array design in your experiment, it is possible to submit only one experiment. If you wish to do this, please contact GEA team.
Microarray data submission
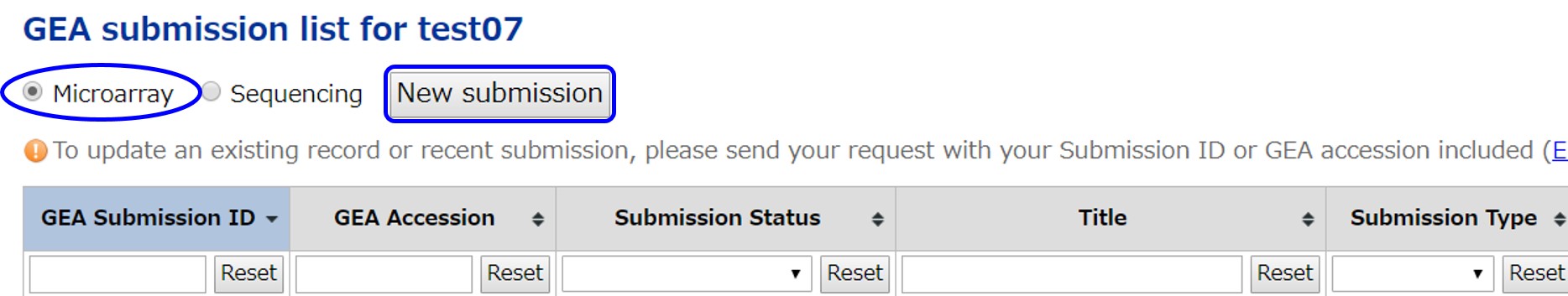
Create a new submission
Login D-way and navigate to the GEA submission page from the top menu.
Create a new microarray experiment submission by selecting “Microarray” and clicking the [New submission]. At the same time, in the DDBJ file server (ftp-private.ddbj.nig.ac.jp), a corresponding subdirectory is created under the submitter’s GEA upload directory. Upload data files to this subdirectory.
List of submission status is as follows. The GEA team reviews submission whose status is in “submission_validated” or “data_error”.
List of submission status
| Status | Explanation |
|---|---|
| New | Metadata are not submitted. |
| Data Submitted | Metadata and data files are submitted. |
| Data Validating | Validating data files. |
| Validation Error | Error occurred in data validation process. |
| Data Validated | Metadata and data are validated. |
| Curating | Curator is reviewing the submission. |
| Accession Issued | Accession number is issued to the submission. |
| Private | Archive files are created and submission is kept private |
| Public | Released to public. |
Upload raw and processed data files
Regarding how to upload your data files, please see “Data upload”.
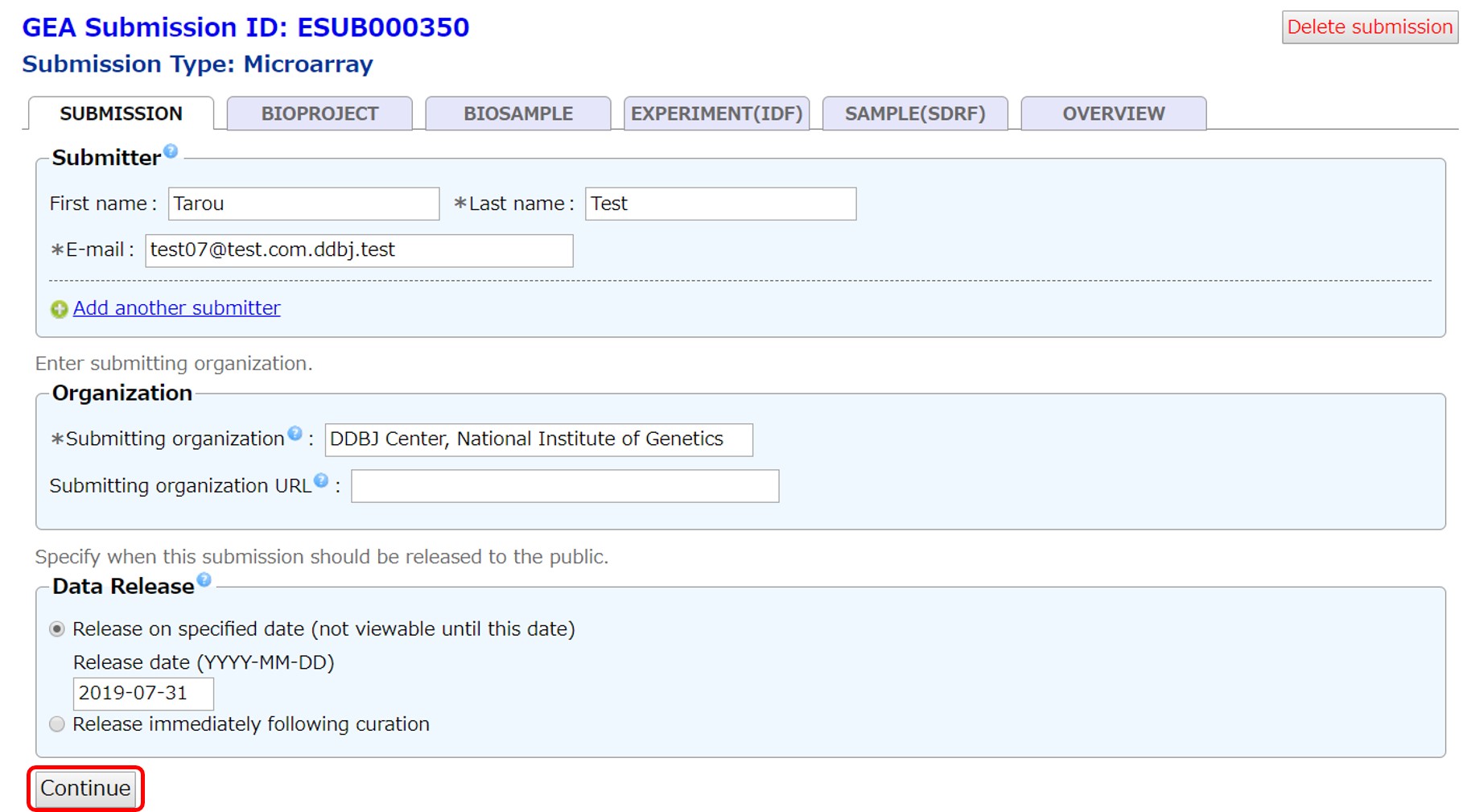
Submission
Set the hold date within four years or choose immediate release when processed. Submitters’ name and affiliation will be public but e-mail address will not be disclosed.
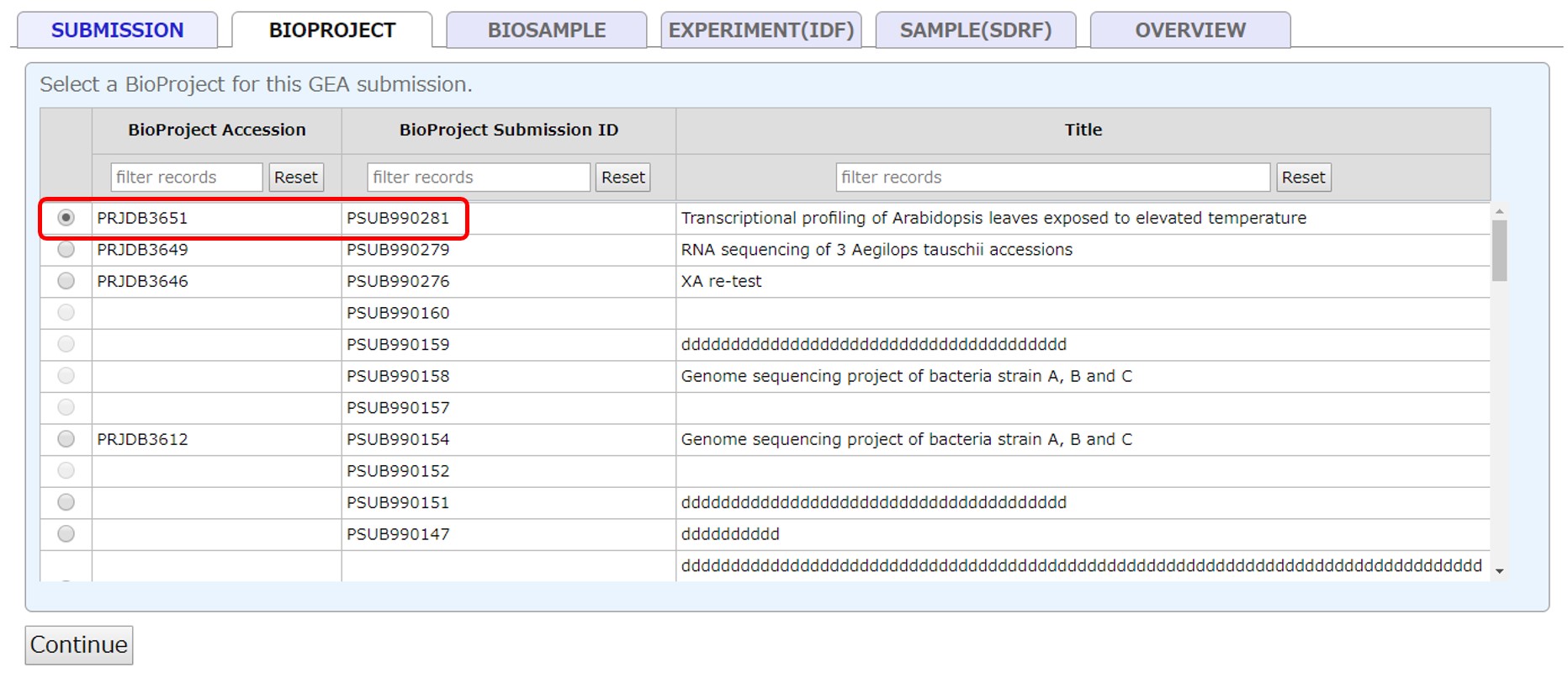
BioProject
Select a submitted project registered in your account. If a BioProject is not registered, please submit a new project at BioProject submission site. To use a project registered in another account, please apply cross-reference.
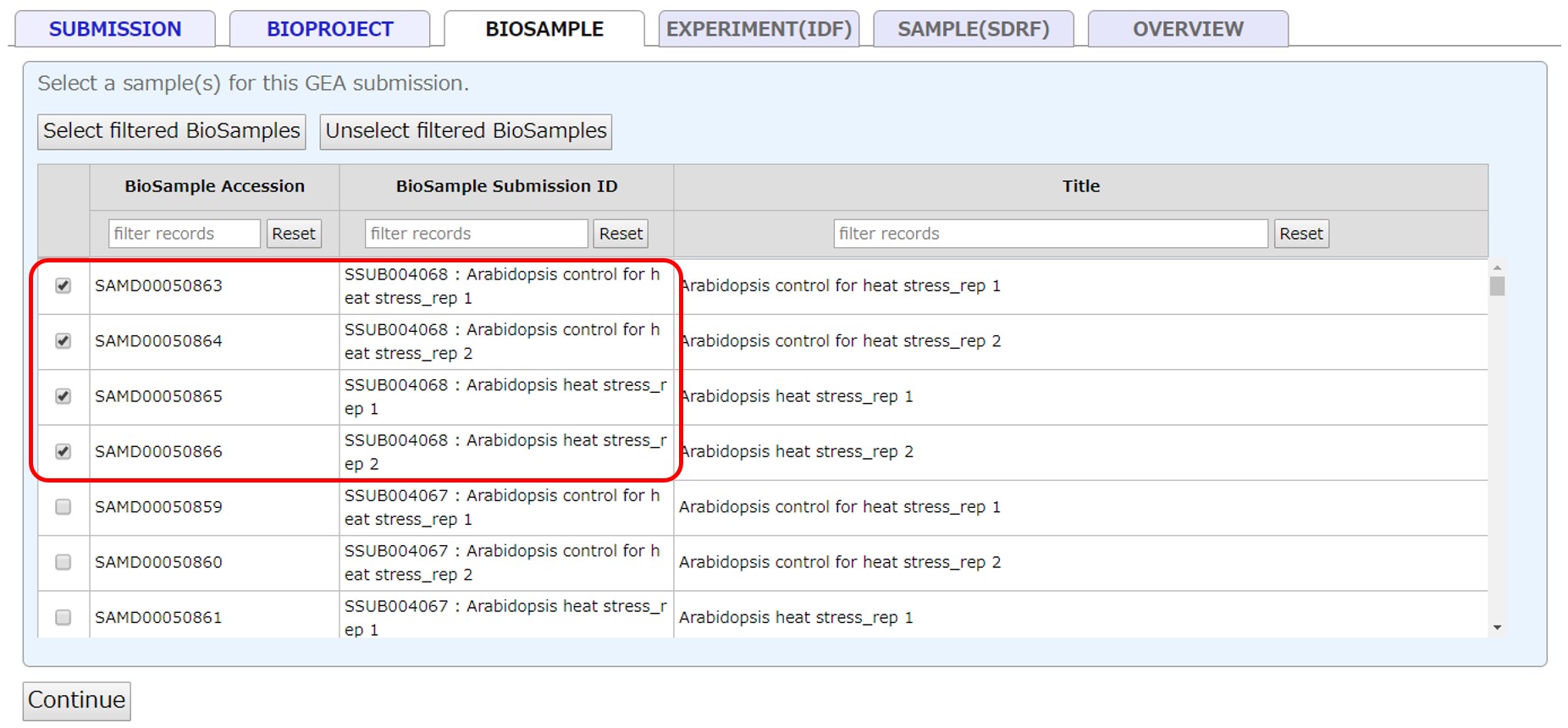
BioSample
Select submitted BioSamples registered in your account. If BioSamples are not registered, please submit new samples at BioSample submission site. To use samples registered in another account, please apply cross-reference.
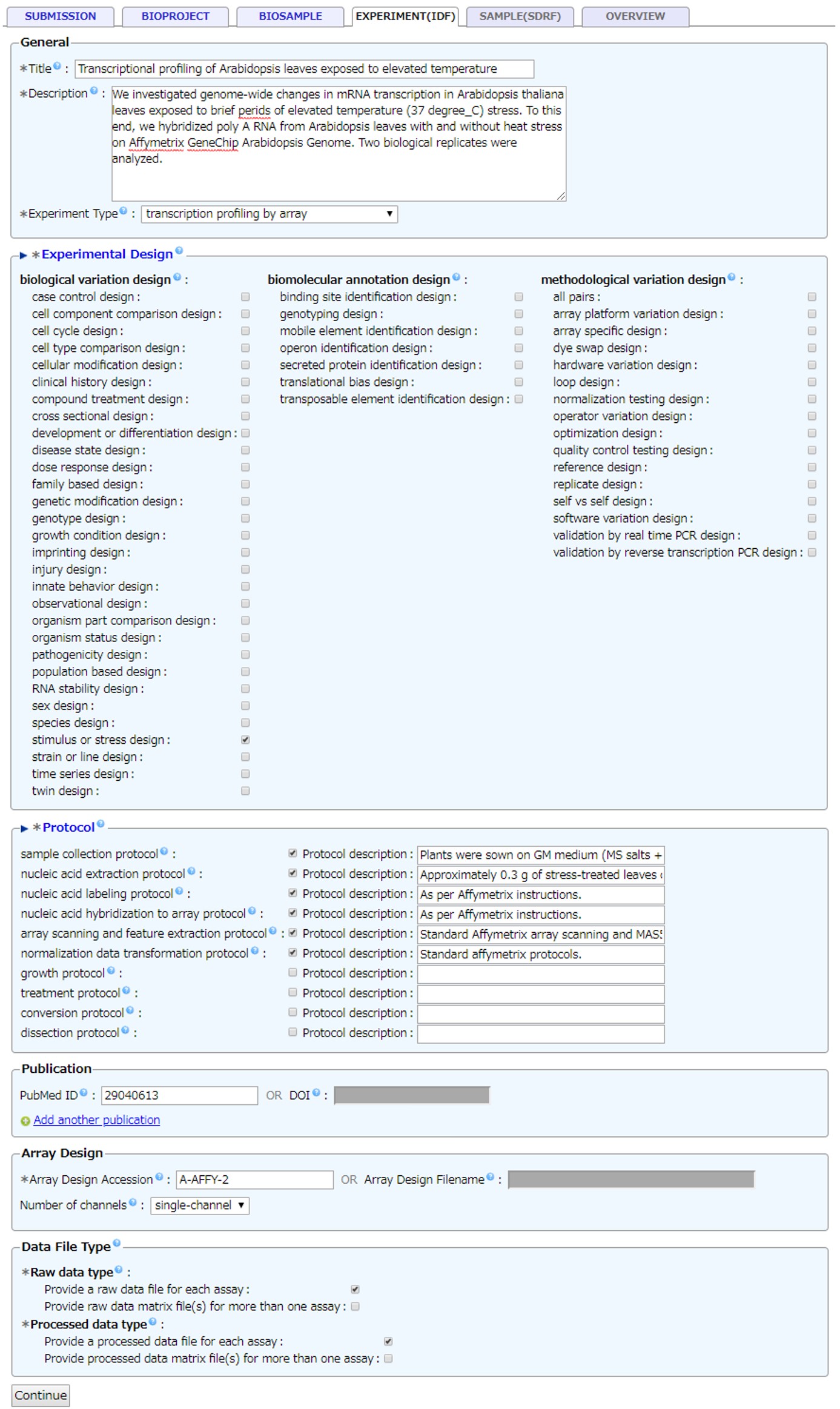
IDF
Enter information for IDF (Investigation Description Format). Example IDF
- Protocol: Pre-checked protocols are mandatory.
- Publication: Describe associated publications by PubMed ID or DOI. For unpublished manuscript, please inform us the publication ID after assignment.
- Array Design: When an array design is available in ArrayExpress/GEA, enter an array design accession number “A-XXXX-n”. When an array design is not available, register a new array design by uploading an array design file into the GEA submission directory.
- Data File Type: Raw and processed data files are required for microarray experiment submission. We strongly recommend to submitting raw and processed data file per sample. Accepted Data Files Formats for microarray experiment.
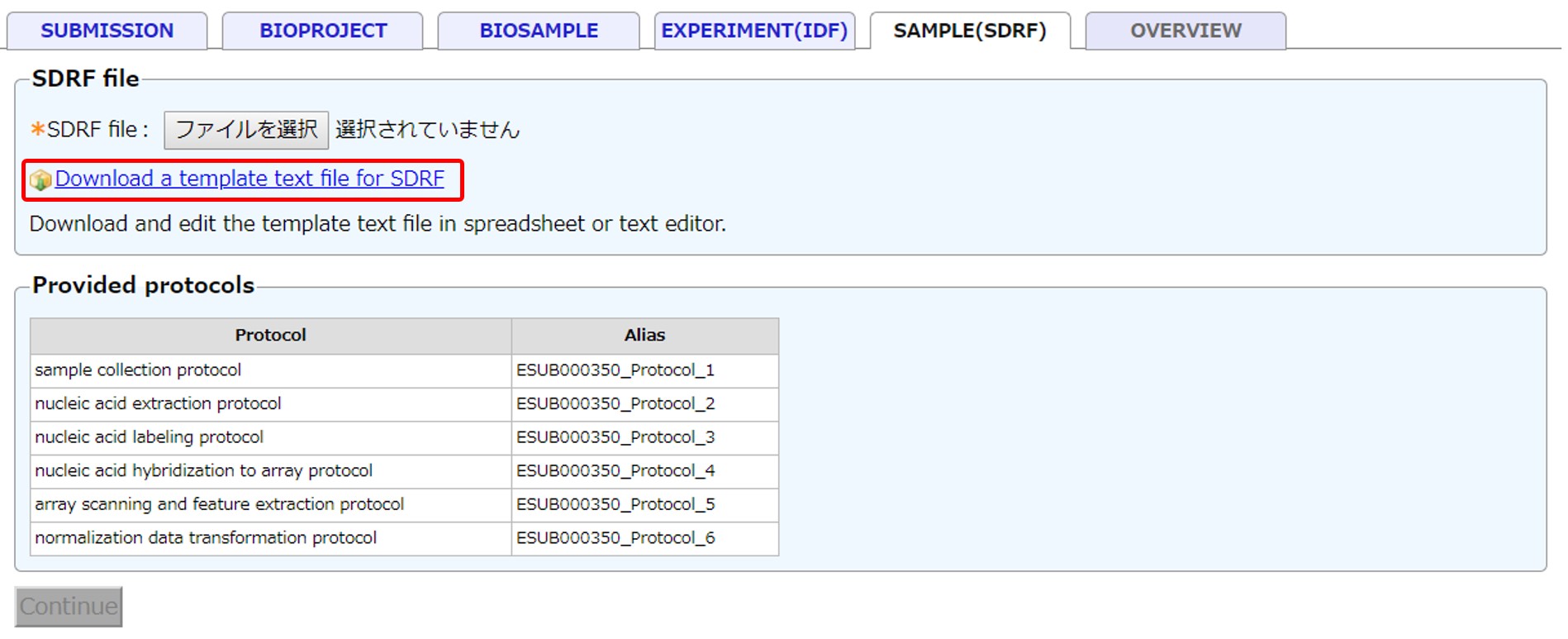
SDRF
Enter information for SDRF (Sample and Data Relationship Format). Example SDRF
Auto-filled fields.
- Name columns and attribute columns for Source Name: Generated from BioSamples.
- SDRF rows: 1 row for 1 BioSample.
- Protocols: Protocols described in IDF are inserted to appropriate positions of SDRF with temporary protocol IDs (e.g., ESUB000350_Protocol_1)
- Technology Type: “array assay” for microarray submission.
- Array Design REF: array design accession or filename described in IDF.
Enter required fields by overwriting <Required: fill in the content> tags.
Fields you need to add.
- Material Type: Enter controlled terms.
- total RNA
- polyA RNA
- cytoplasmic RNA
- nuclear RNA
- genomic DNA
- protein
- other
- Label: Enter label compounds used to label the extracted molecule such as biotin, Cy3 and Cy5.
- Array Data File and Comment[Array Data File md5]: Enter filename and md5 checksum pair for each raw data file.
- Derived Array Data File and Comment[Derived Array Data File md5]: Enter filename and md5 checksum pair for each processed data file.
- A list of filename and its md5 checksum (output of md5sum command) can be provided as a file
.md5 (e.g., ESUB000001.md5) (when the checksum values are provided in both SDRF and .md5 file, those in the .md5 are used). - Factor Value[enter experiment factor name here]: A user-defined name for each experimental factor studied by the experiment. These experimental factors represent the variables within the investigation (e.g. growth condition, genotype, organism part). The actual values of these variables will be listed in the “Factor Value []” columns.
Example: - Factor Value[strain]
- AT76
- KU-2003
- KU-PI499262
Select the entered SDRF file and continue.
md5 checksum
GEA uses md5 checksum values to detect corruption of files.
When there are many files or you are familiar with command line, provide output of md5sum command as a separate file with the filename
Example: ESUB000001.md5 (checksum values and filenames are delimited by two spaces)
ed3d9b2adb5b29aa476b9d4164e208d5 raw1.txt
3d77463ca6f43416a6c1925b7704d304 raw2.txt
0e5be28700daa6d61ea3351921d6e578 processed1.txt
351fb1324feebe954405ca610e46ae44 processed2.txt
3d5749b63617da9002c7376deee8e0a3 array-design.txt
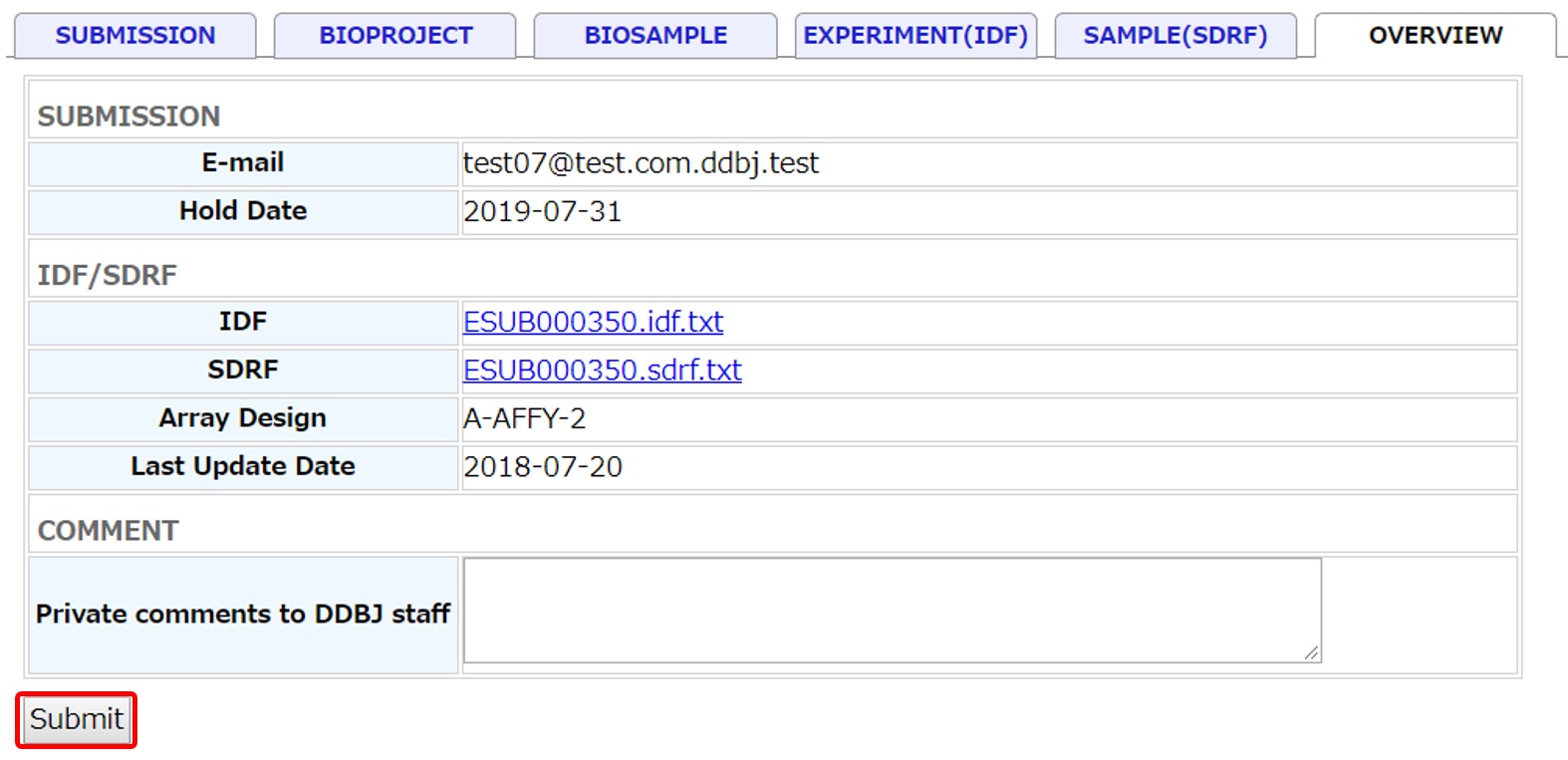
Overview and submit
You can download the IDF and SDRF files and check them. When correction is necessary, go back to the previous tab and corrent metadata.
Submit the IDF and SDRF metadata by clicking [Submit].
Validation
When data files described in the IDF and SDRF are not found in the submission directory, an error message “Data file is not uploaded” is shown and the submission is aborted.
The validator checks submitted IDF and SDRF files according to the validation rules and gives warning and error messages. Errors need to be resolved for submission.
Accession number
GEA accession numbers are issued to completed GEA experiment. You can allow reviewers access to private records by communicating a reviewer accesss token.
Update submission
For updates or deletions, please request them via the form to the GEA team.