BioSample
BioSample Submission
- Submission of research data from human subjects
- For submitting data from human subjects (human data) to the databases of DDBJ center, it is submitter’s responsibility to ensure that the dignity and right of human subject are protected in accordance with all applicable laws, ordinances, guidelines and policies of submitter’s institution. In principle, make sure to remove any direct personal identifiers of human subjects from your data to be submitted. Before submitting human data, read the “Submission of research data from human subjects”.
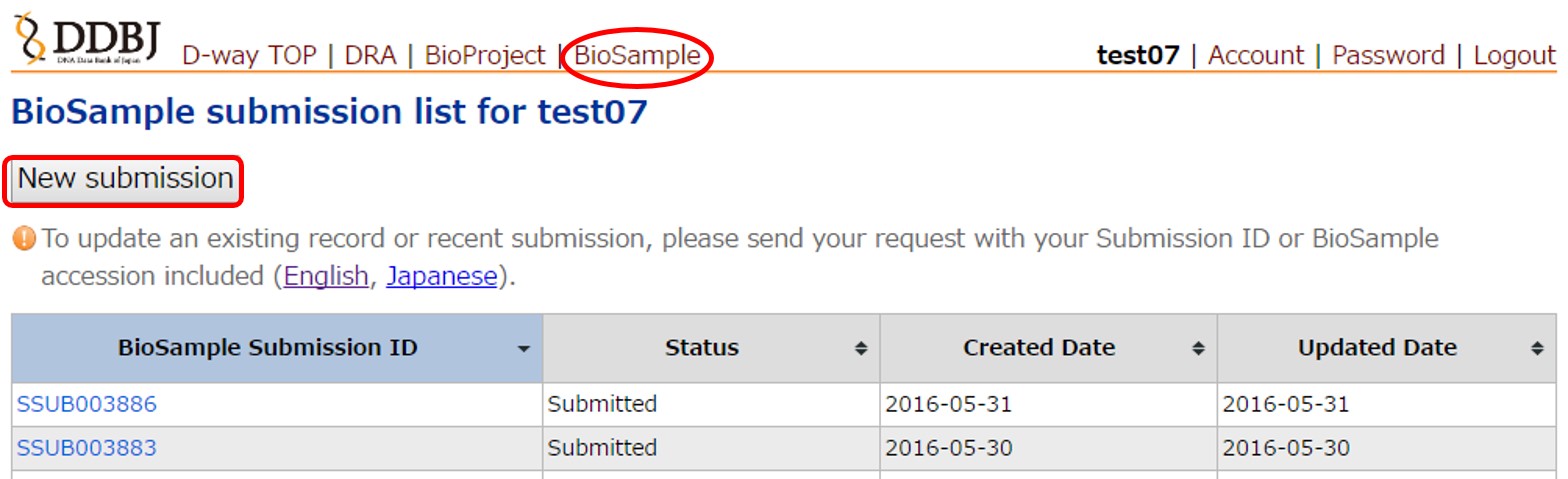
Create a new sample submission
Obtain a submission account.
Move to the Biosample submission page from the “Biosample” menu at the top. Create a new sample submission by clicking the [New submission] button.
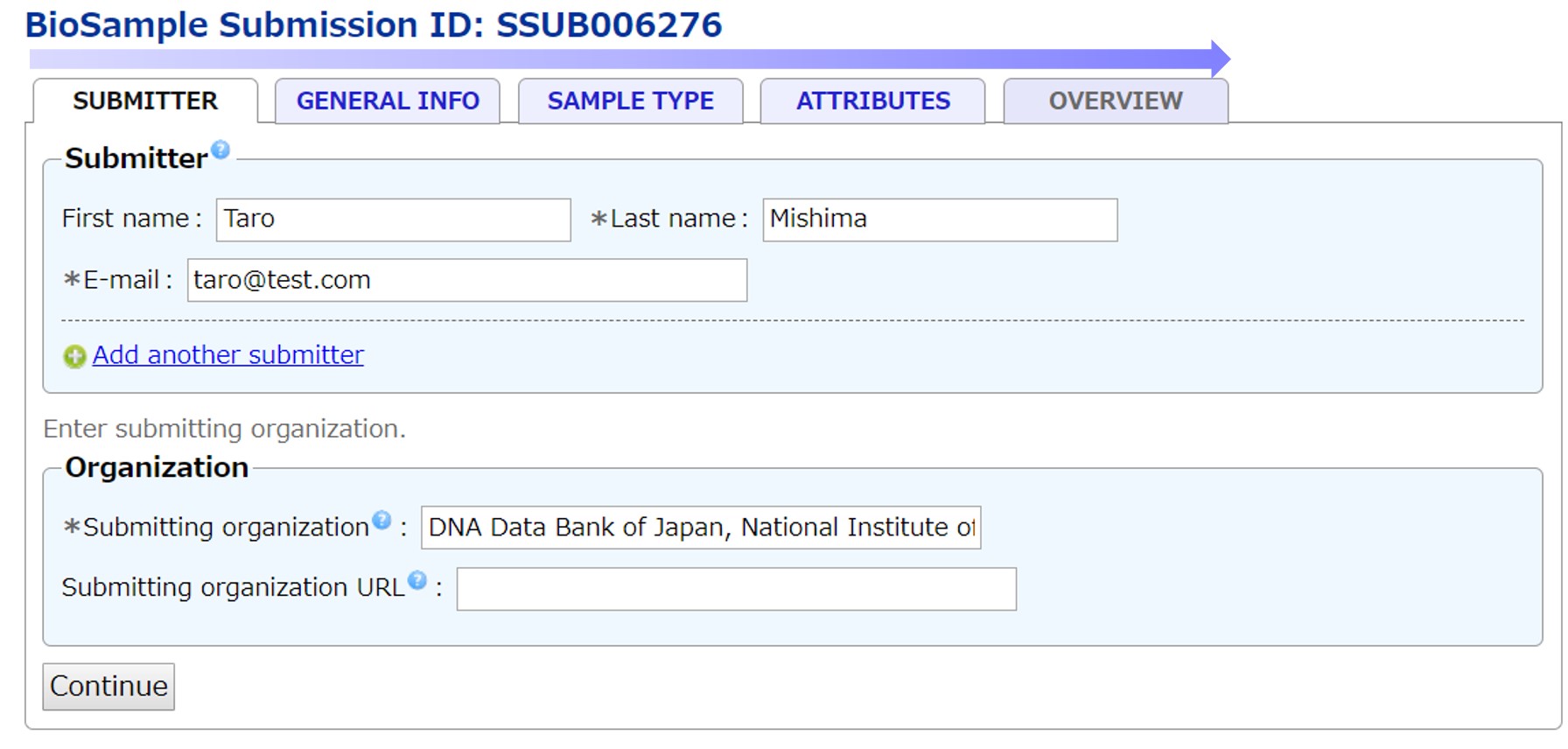
To submit a BioSample, enter content from left to right tabs.
For BioSample information fields, please see the BioSample information fields.
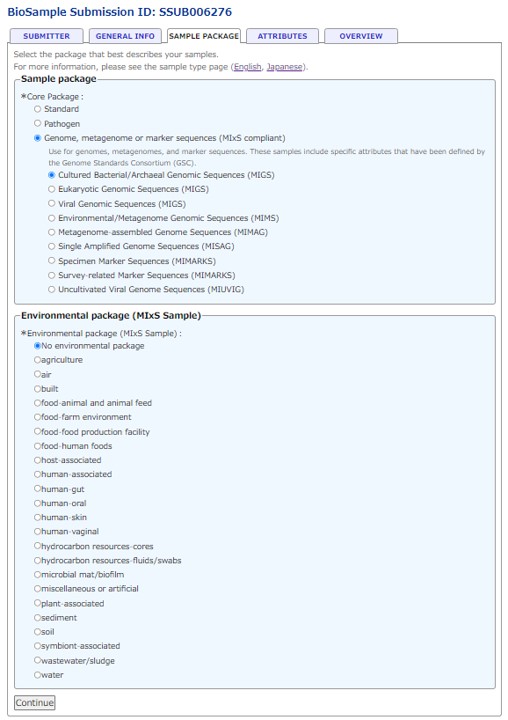
Select a sample type in the “SAMPLE TYPE”.
In the “SAMPLE TYPE”, select an appropriate package according to the type of sample or sequence. Enter required and optional sample attributes provided by a selected package.
- See “Sample Information” regarding how to select a package.
- See “Sample attributes” regarding sample attributes.
- More than one sample can be submitted in single submission.
- Single submission can not have samples of different packages.
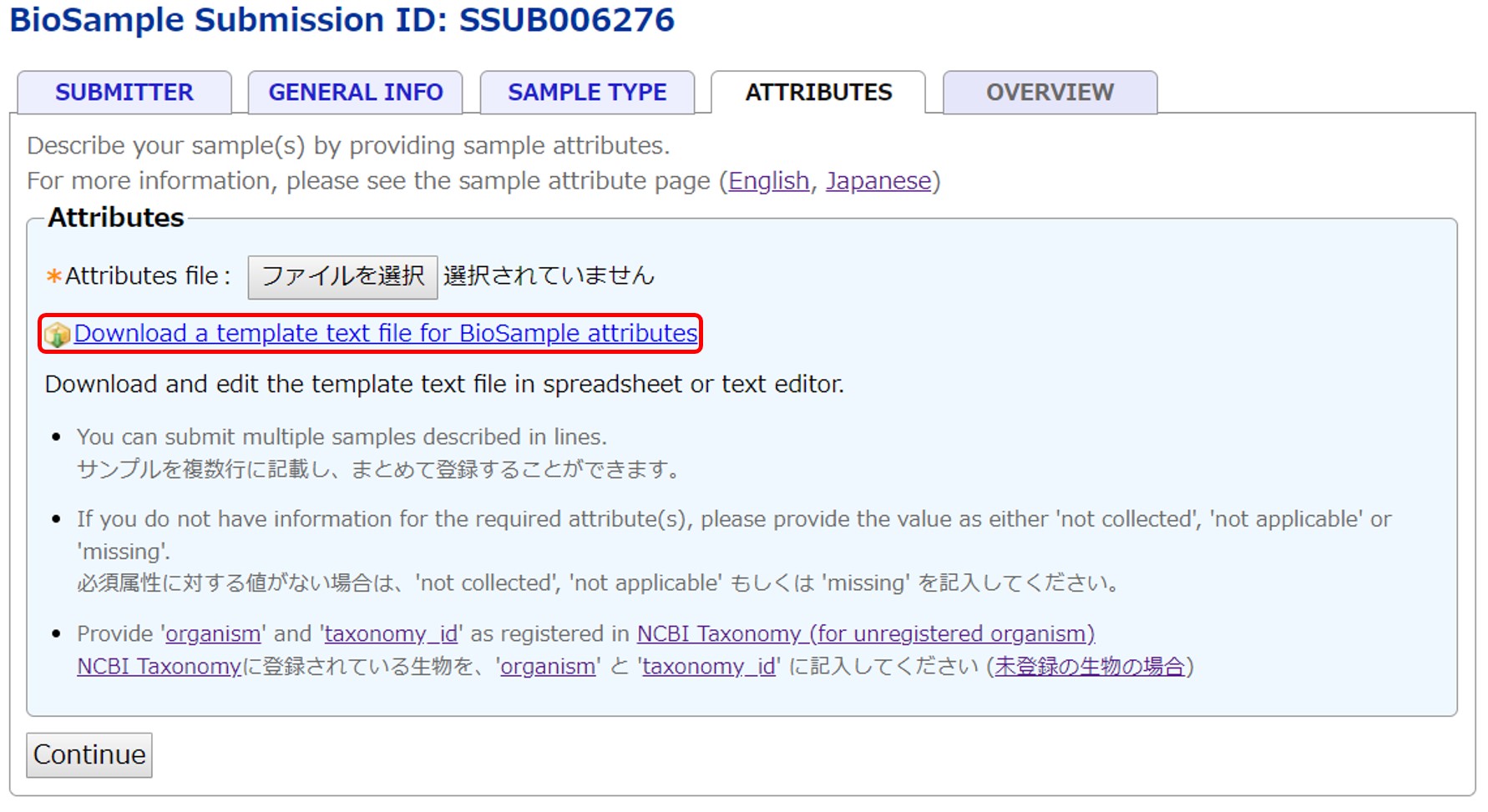
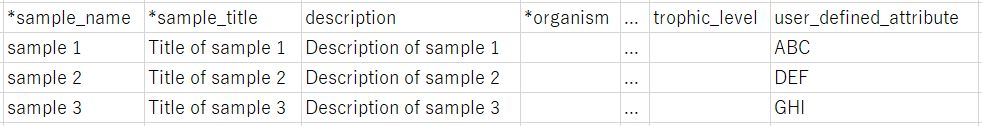
Download a template text file according to the selected package. User-defined attributes can be added to the rightmost column.
Enter sample attributes
A text file is separated by tab and can be opened and editted in spreadsheet editor (e.g. Excel®). Attribute names are in a header line. Attributes with “*” are required.
From second lines, enter one sample per line.
Missing value reporting
The International Nucleotide Database Collaboration (INSDC) have developed a standardised missing/null value reporting language to be used where a value of an expected format for sample information reporting can not be provided. Submitters are strongly encouraged to always provide true values of expected formats. In cases where this information cannot be provided (e.g., pathogen samples for which this information would lead to identifiability of a human) or is not relevant (e.g., study of a model organism lab stock or an established cell line), it is recommended to declare an appropriate exemption using one of the reporting level terms of the extended INSDC “missing value” reporting standards (e.g. “missing: control sample”). If these reporting level terms are not appropriate, enter “missing”, “not collected” or “not provided”. The repoting level terms are required for two mandatory attributes “collection_date” and “geo_loc_name”.
To facilitate an understanding of the supported terms we enclose a table with the missing/null value terms and their definitions.
Please use the following standardised missing value vocabulary only if a true value of an expected format for a mandatory field is missing. If a true value is missing for a recommended or an optional field then these fields should not be used for reporting at all.
INSDC missing value reporting terms (INSDC website)
| INSDC term (top level) | INSDC term (lower level) | Definition | INSDC term (reporting level) | Definition |
|---|---|---|---|---|
| not applicable | information is inappropriate to report, can indicate that the standard itself fails to model or represent the information appropriately | control sample | Information is not applicable as the sample represents a negative control sample collected in a lab. | |
| sample group | Information is not applicable as the sample represents a group of samples that do not have a single origin. E.g. for co-assembly or transcriptome assembly. | |||
| missing | not collected | information of an expected format was not given because it has not been collected | synthetic construct | Information does not exist as the sample represents an ab-initio synthetic construct. |
| lab stock | Information was not collected as the sample represents a cultured cell line or model organism under long-term lab control. | |||
| third party data | Information does not exist as the metadata was not collected or reported in records predating the 2023 agreement. For use in Third PArty data submissions. | |||
| not provided | information of an expected format was not given, a value may be given at the later stage | data agreement established pre-2023 | Data agreements were established before the 2023 INSDC standard and metadata can not be provided. A value may be given at a later stage. | |
| restricted access | information exists but can not be released openly because of privacy concerns | endangered species | Information can not be reported as the target organism is endangered e.g. on the IUCN red-list. | |
| human-identifiable | Information can not be reported as the metadata would make the sample human-identifiable. |
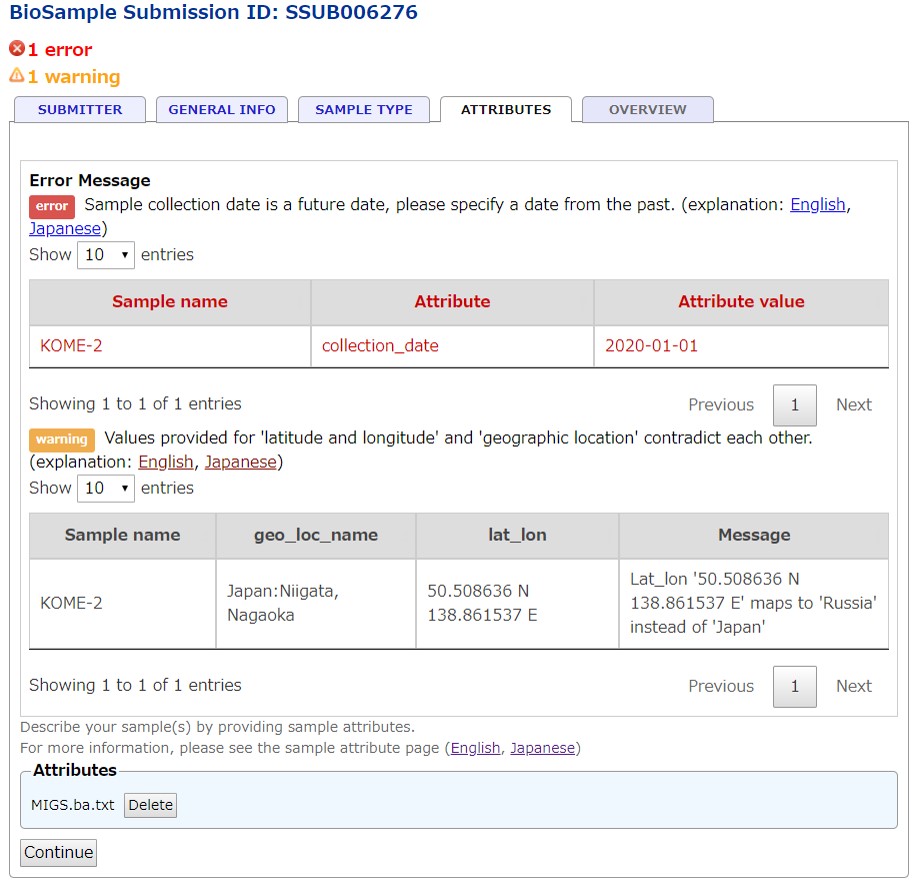
Sample attributes validation
Upload the sample attribute file by selecting the file and click the Continue button. The validator checks the uploaded file accoring to the rules and feedbacks the error and warning messages. Submitters can not submit the BioSample unless all errors are resolved.
For validation rules and messages, please see Validation rules page.
In the following packages, at least one of attributes is required. For example, strain or isolate is mandatory in the Microbe package.
In the BioSample submission tsv, required attributes are marked with “*”, however, at least one required attributes are not marked.
| Package | ‘either-one-mandatory’ attributes | ‘either-one-mandatory’ attributes |
|---|---|---|
| Microbe | strain, isolate | isolation_source, host |
| Model.organism.animal | strain, isolate, breed, cultivar, ecotype | age, dev_stage |
| Metagenome.environmental | isolation_source, host | |
| Invertebrate | isolate, breed | isolation_source, host |
| Plant | isolate, cultivar, ecotype | age, dev_stage |
| Virus | host, lab_host | |
| Beta-lactamase | strain, isolate | |
| Pathogen.cl | strain, isolate | |
| Pathogen.env | strain, isolate |
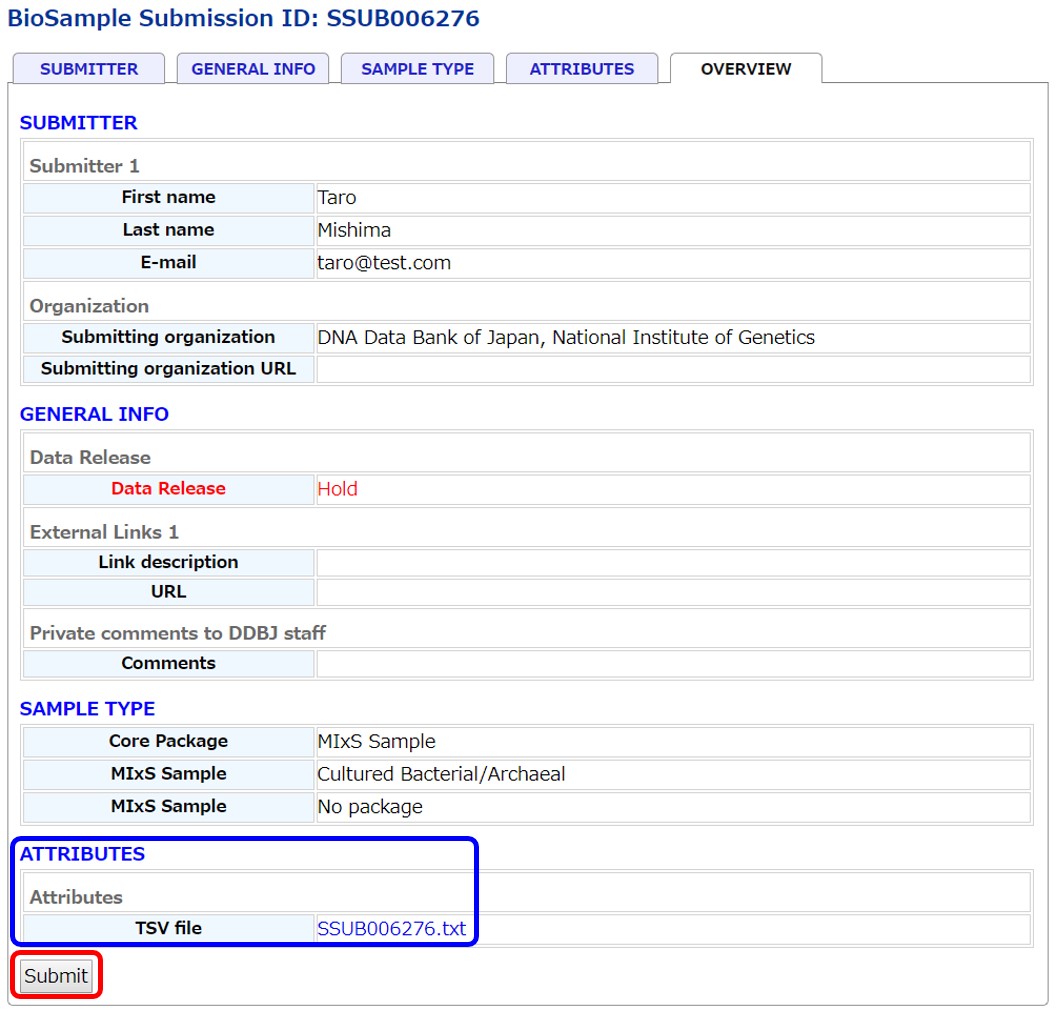
Check content in the last “OVERVIEW” and submit samples. In the “ATTRIBUTES” area, the submitted sample attribute file can be downloaded.
Accession numbers
When creating a new submission, a temporary ID starts with SSUB is assigned. The DDBJ BioSample issues accession numbers with prefix SAMD to the submitted samples passed validation. When an unregistered organism is described in the organism or the locus_tag_prefix has values, accession numbers are issued after curator review. You can view status and accession numbers of submitted samples in D-way.
Release of BioSample
You can select the following options. Hold date cannot be set for BioSample.
- Release immediately following curation
- Release when referenced data is published
The submitted sample data can be kept private. Sample data are automatically released when the linked DDBJ/DRA/GEA/MetaboBank record(s) is published. The release of the BioSample record does not trigger the release of private DDBJ/DRA/GEA/MetaboBank record(s) referencing this BioSample accession. The release of the BioSample record does trigger the release of referencing BioSample in derived_from attributes.
FAQ: How are linked BioProject/BioSample/sequence data released?
Update BioSample
Registered samples can be updated. Please inform us points to be updated through the BioProject/BioSample/DRA update request form so that we will update the samples. If you want to update sample attributes, please attach the updated attribute tsv file to the email replying to the accession number notification email. You can download the attribute tsv file at D-way.