BioProject
BioProject Submission
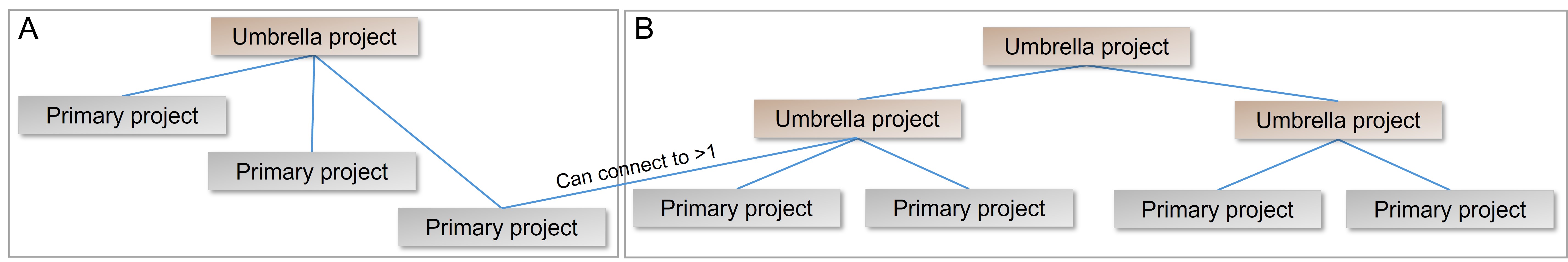
Primary and umbrella projects
There are two types of project: primary and umbrella.
- Primary project:
- Submitted projects which are intended to represent and be linked to current or future data submissions. Primary projects can be kept private.
- Umbrella project:
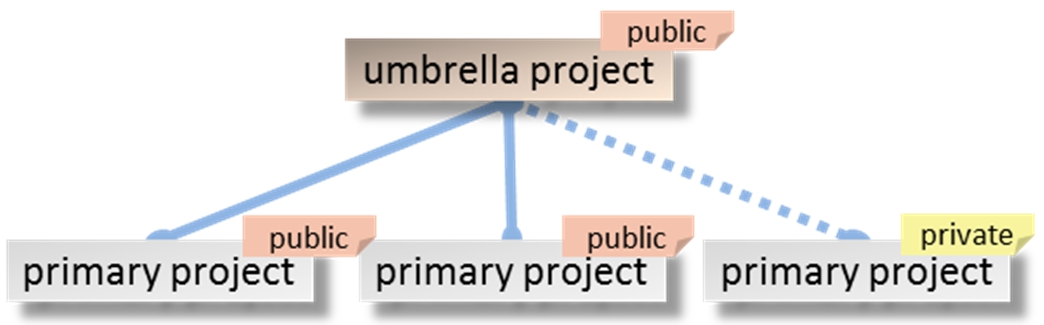
- Administrative project that is created to group multiple projects that are related by a single effort from a single submitter or group of submitters. Umbrella projects cannot be kept private.
- Submitted data can directly refer to primary projects but can not refer to umbrella projects. The data are indirectly linked to the umbrella project through the primary project.
- Submitted primary projects are not directly linked to other primary projects: they are linked indirectly by way of links to the umbrella project.
Hierarchy
Some large initiatives are represented by more than one layer of umbrella projects (see Figure B below), for instance, a top-most level may identify the largest definition of the collaboration, a second level of umbrella projects represent sub-projects at participating institutions, and finally a third layer represents the projects that actually generate the data that is submitted. Primary and umbrella projects can be associated with more than one umbrella project.
Cases requiring project registration
Please check whether a project registration is necessary for your data submission atsubmission navigation.
Submit a new BioProject submission
Obtain a submission account.
Submit a new project by clicking the [New submission].
To submit a BioProject, enter content from left to right tabs. Submitter information is copied from the account information. For BioProject information fields, please see BioProject information.
To submit genome sequences with annotations to DDBJ, locus tag prefix(s) should be registered in BioSample.
Check the content in “OVERVIEW” and submit a project from [Submit].
Accession number
A BioProject accession number with prefix PRJDB is automatically assigned to the submitted project.
When the data release is specified as “Release immediately following curation”, the project will be automatically released at night.
- Do NOT cite submission ID with prefix PSUB in your publication.
- Do not double submit the projects to us which have been registered to EBI and NCBI.
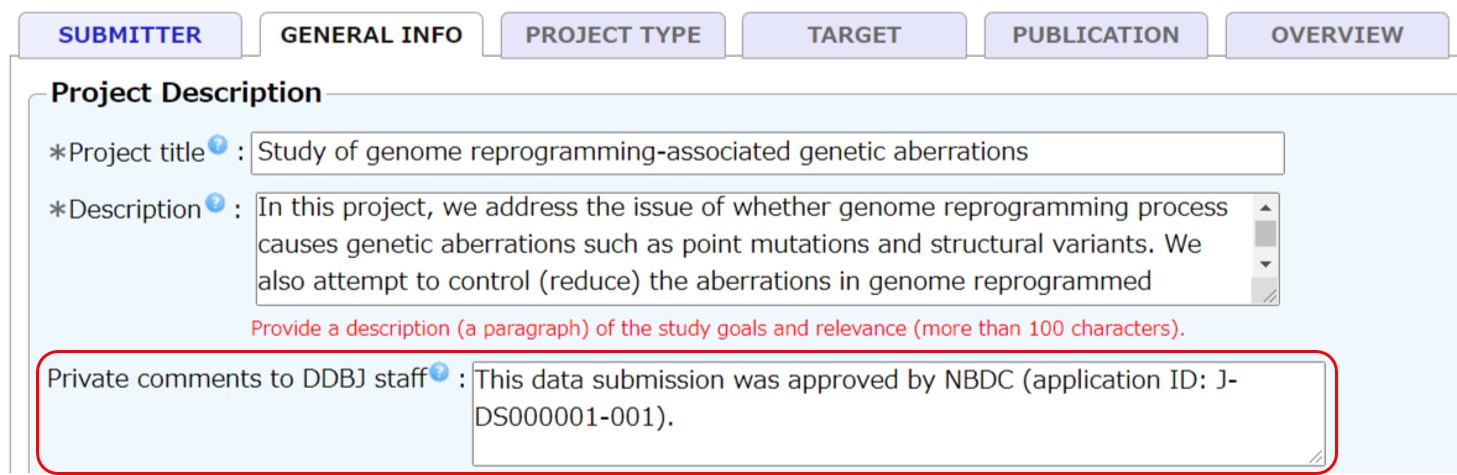
Human data
To submit human data, your data submission application needs to be approved by DBCLS before submission. Write the approved application ID (for example, J-DS000001-001) in “Private comments to DDBJ staff” of BioProject.
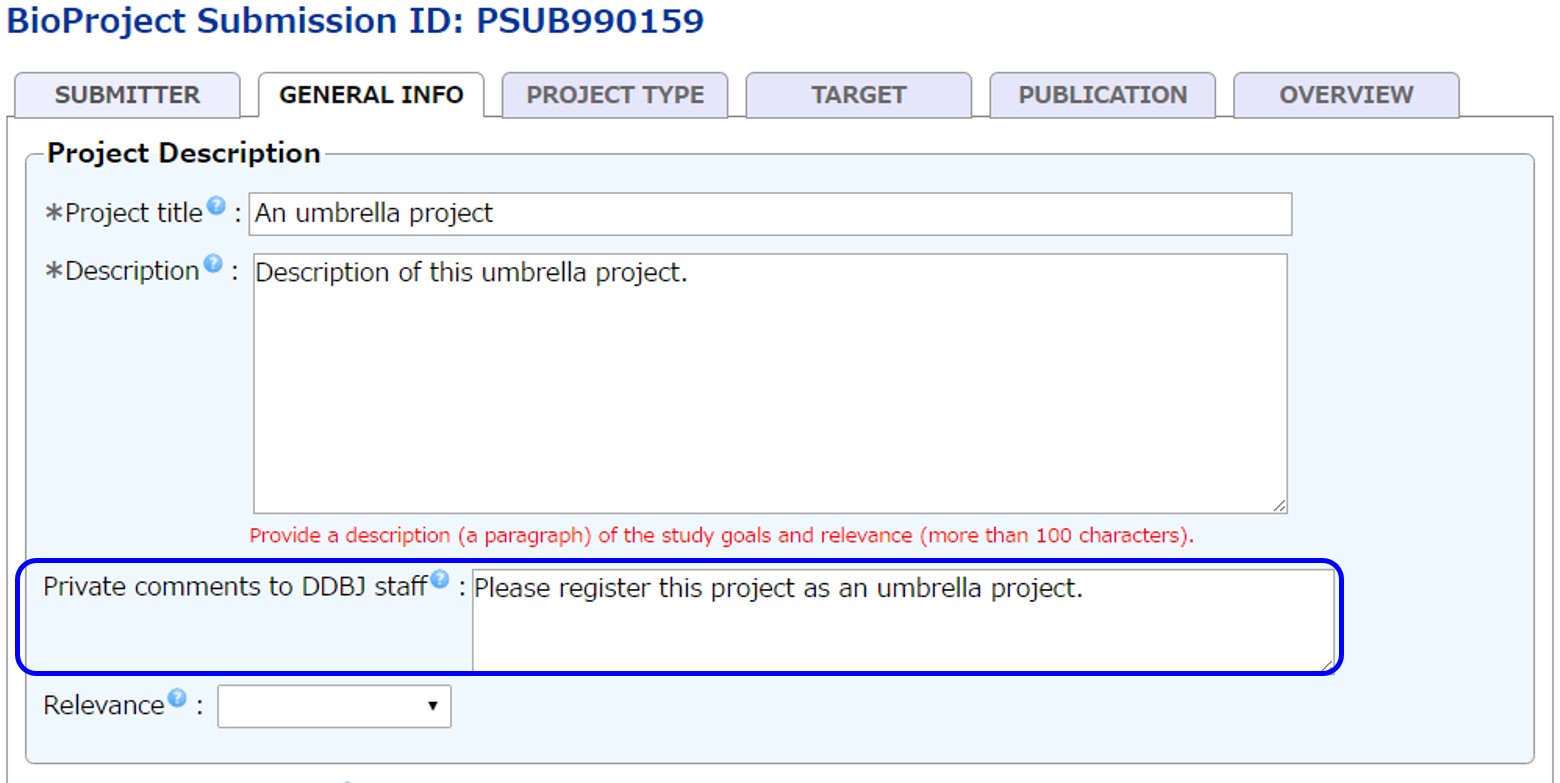
Submit an umbrella project
Umbrella project can be submitted as primary project in D-way. Be sure to tell the DDBJ BioProject staff that submitting project is umbrella by writing so in the Private comments to DDBJ staff. Umbrell project cannot be kept private.
When submitting an umbrella project to organize haplotype datasets, enter accession numbers of primary BioProjects to be linked and their names for haplotypes (e.g. PRJDB1 Principal, PRJDB2 Alternate and PRJDB3 DRA).
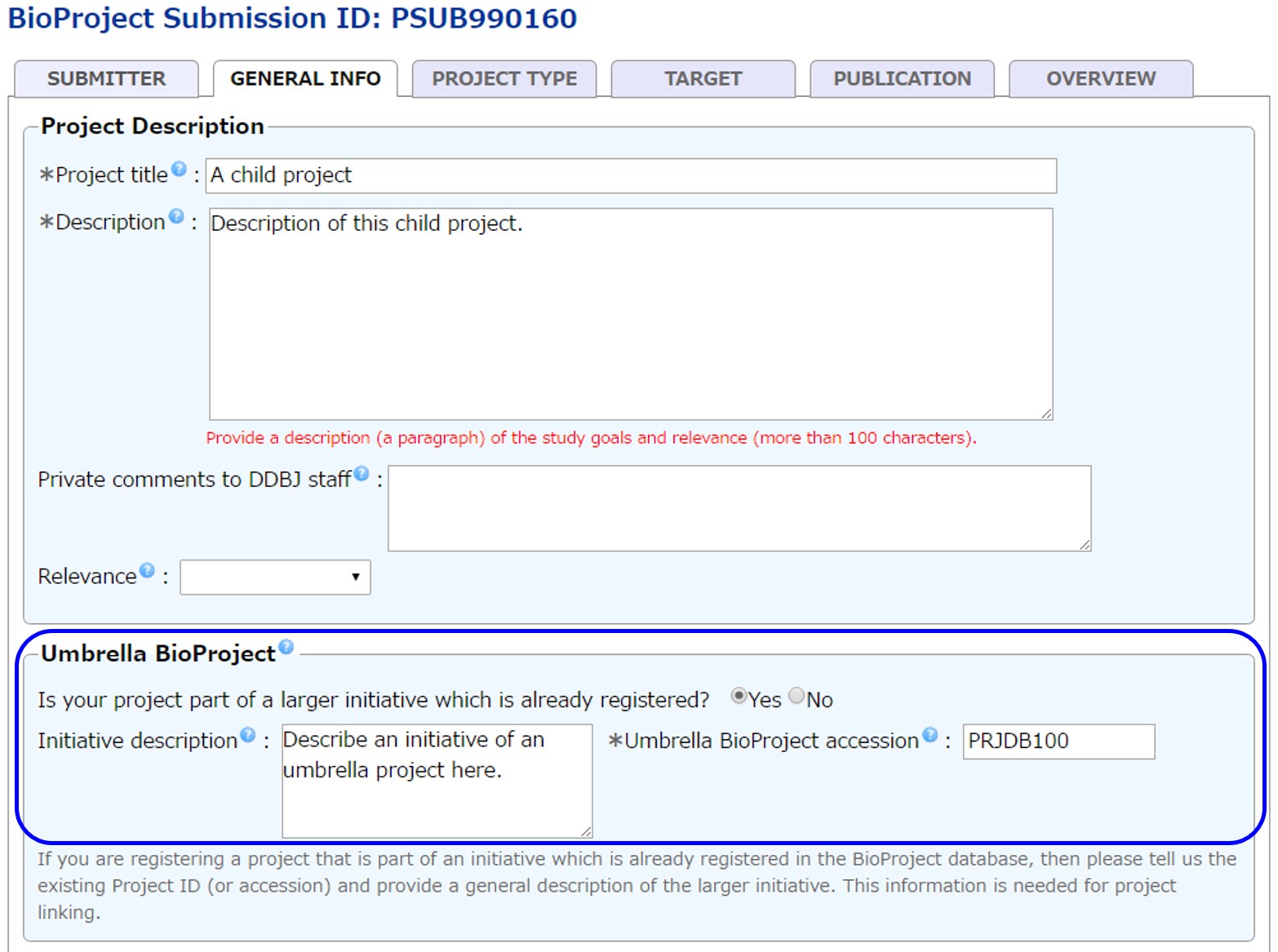
Link primary project to umbrella
When submitting project, in the Umbrella BioProject, enter abstract and accession number of an umbrella to be linked. The DDBJ BioProject staff will link the submitted primary project to the umbrella based on this information.
Release of projects
Registered projects can be released in the following two ways. Hold date cannot be set for BioProject.
- Release immediately after registration.
- Release when records citing the BioProject accession are made public.
The projects can be kept private and are released as follows.
- You can “immediately release” or “hold” the registered primary project.
- The submitted primary project data can be kept private until the linked DDBJ/DRA/GEA/MetaboBank data made be public.
- Hold date of the project data cannot be specified.
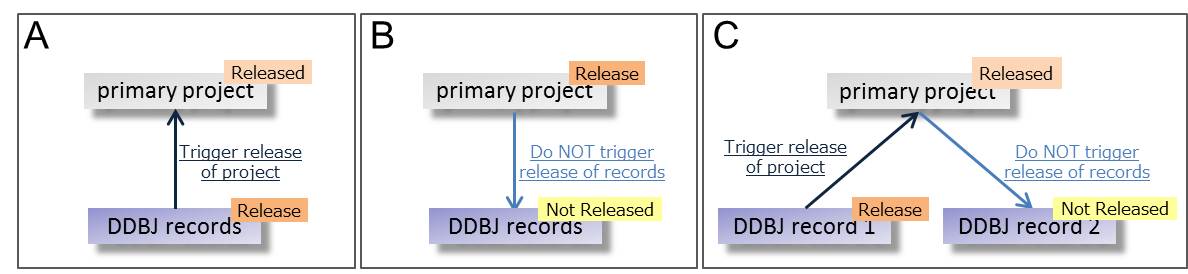
- Primary project data are automatically released when the linked DDBJ/DRA/GEA/MetaboBank data are published.
- Publication of the primary project do not cause automatic release of the linked DDBJ/DRA/GEA/MetaboBank data.
- Under a primary project, publication of data does not cause the indirect release of the other data belong to the same project.
FAQ: How are linked BioProject/BioSample/experimental data released?
An umbrella project cannot be kept private.
An umbrella project can have public and private primary projects. Hierarchical relationship between the public umbrella project and the un-released primary project is invisible.
Released projects are exchanged with NCBIand EBI.
Update
Registered projects can be updated such as addition of publication information. Please request update through BioProject/BioSample/DRA update request form.