DDBJ Annotated/Assembled Sequences
Haplotype
Historically, whole-genome sequencing generated a single consensus sequence without distinguishing between alleles on homologous chromosomes. Long-read sequencing technologies can identify haploid chromosomes. Because two genome sequences are produced from single sample in haplotype sequencing, INSDC establishes the guideline for haplotype sequence submission. At DDBJ we had used the term ‘pseudohaplotype’ for all of these but we use ‘haplotype’ now.
Haplotype submission
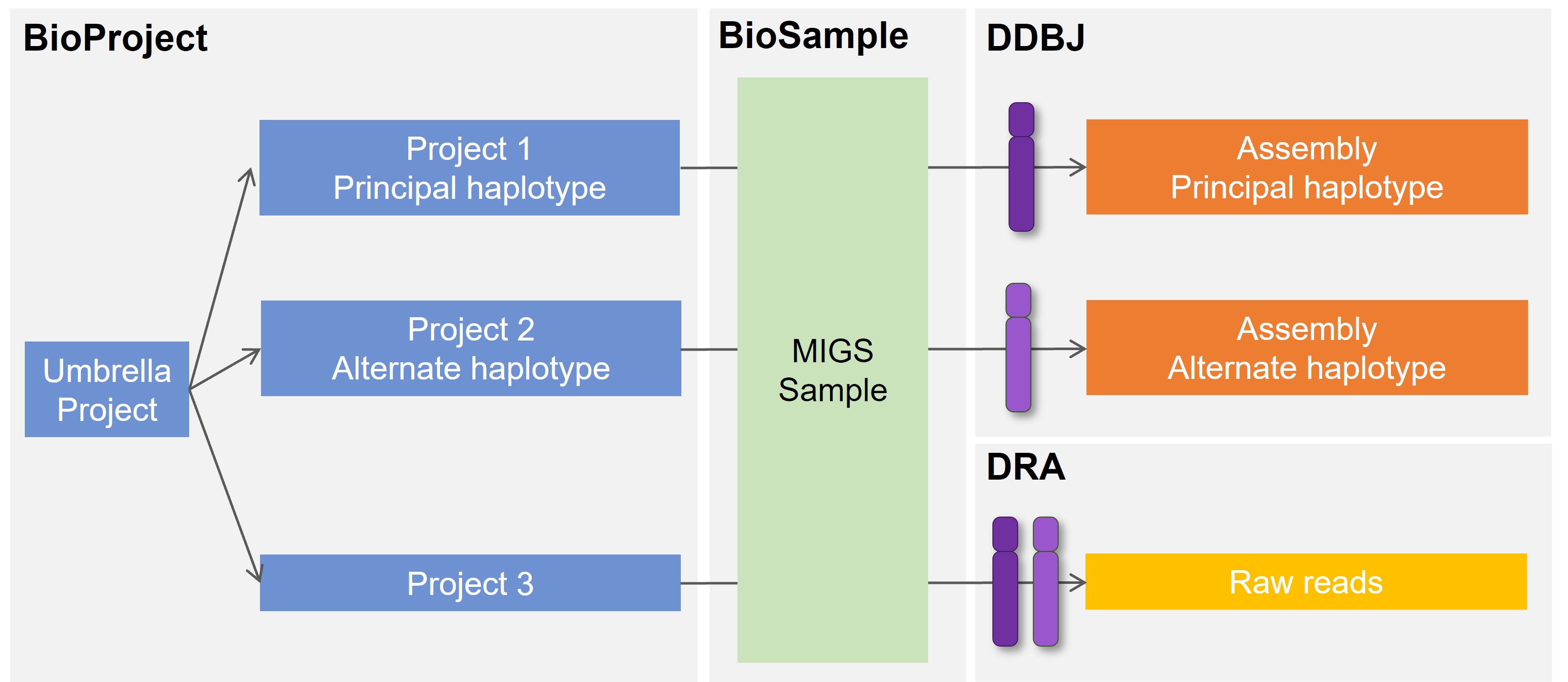
This page describes a typical case of the haplotype sequence submission. To distinguish haplotype assemblies, as an example, name one of the assemblies as “Principal” and another as “Alternate” here. Because each haplotype assembly is derived from the same sample, both assemblies share the same BioSample. Because INSDC manages a genome assembly by unique combination of BioProject and BioSample, create separate BioProject for each principal and alternate haplotype to make the combination unique. Create an umbrella BioProject to group these projects.
If the raw DRA sequencing data contain reads from both haplotypes, create a BioProject for DRA apart from those for assemblies. If the DRA data are derived from the same sample for the assemblies, use the same BioSample.
When more than one haplotype dataset exist (e.g. three haplotype datasets of species A, B and C), create a common umbrella project and group each primary projects.
Naming haplotype assemblies
There are a few naming options to distinguish haplotype assemblies. One of the options must be asserted by the submitter.
- Principal haplotype/Alternate haplotype: if one is much better (Principal) than the other (Alternate).
- Haplotype 1/Haplotype 2: if they are of similar quality. When more than 2 haplotypes are present, increase the number like Haplotype 3/Haplotype 4.
- Maternal haplotype/Paternal haplotype: when that information is known.
BioProject
Create separate BioProject for each principal and alternate haplotype and an umbrella BioProject to group these projects. When submitting the umbrella project, enter accession numbers of primary BioProjects to be linked and their names for haplotypes (e.g. PRJDB1 Principal, PRJDB2 Alternate and PRJDB3 DRA).
- BioProject 1: Principal
- Add phasing information in the title. For example, Principal haplotype or Primary haplotype.
- BioProject 2: Alternate
- Add phasing information in the title. For example, Alternate haplotype or Alternate haplotype.
- Umbrella BioProject
- For grouping BioProject 1, 2 and the other related BioProjects (BioProject 3 for DRA in the figure).
BioSample
Because the sample is shared by haplotypes, create single BioSample.
- Select MIGS package.
- Create a common BioSample for principal and alternate haplotype.
- If you add gene annotations to the haplotype sequences, enter a locus tag prefix you want to use for the principal and the alternate haplotype in the locus_tag_prefix attribute. The locus tag prefix is shared by the principal and the alternate haplotypes, loci can be distinguished by tags, for example, A1C_p00001 (principal) and A1C_a00001 (alternate).
DDBJ
Submit the principal and the alternate haplotype sequences.
- Principal haplotype
- Reference the BioProject 1 (Principal) in DBLINK.
- Add the pre-defined comment in ST_COMMENT. Genome-Assembly-Data ST_COMMENT: Diploid :: Principal Haplotype
- Alternate haplotype
- Reference the BioProject 2 (Alternate) in DBLINK.
- Add the pre-defined comment in ST_COMMENT. Genome-Assembly-Data ST_COMMENT: Diploid :: Alternate Haplotype
Real-world examples
Common
- BioProject: PRJDB10054 (Umbrella)
- BioSample: SAMD00229903
Principal haplotype
- BioProject: PRJDB10055
- DDBJ: BLYA01000001-BLYA01003780
Alternate haplotype
- BioProject: PRJDB10056
- DDBJ: BLYB01000001-BLYB01003780
DRA
- BioProject: PRJDB9979
- DRA: DRR231909-DRR231923