DDBJ Annotated/Assembled Sequences
Organism qualifier
Every value of /organism qualifier must be
- a “scientific name” (not svnonym, equivalent name or others)
- ranked as species or lower taxa (subspecies, variety, strain, etc.)
taxonomy database
All organisms that are represented in the sequence data of DDBJ are registered to
the taxonomy database.
For construction of the nucleotide sequence database, it is important to manage the organism names
for the data and also necessary to unify the diverse names of the organisms.
The taxonomy database is used as the reference database for the unified organism names.
The primary purpose of the taxonomy database is to unify descriptions of organism names.
Consequently, the taxonomy database is not an authoritative source for nomenclature or classification.
A taxonomic name may differ from the submitter’s proposal or widely used taxonomic name because only the organism name in the taxonomy database
which is managed by GenBank can be used for the entry.
Please refer to description of the taxonomy database.
DDBJ provides a web service called
TXSearch to retrieve organism names in the taxonomy database.
This would be helpful as a reference of taxonomic names when you submit nucleotide sequences to DDBJ.
Quick guide to describe organism names
In cases of the organism names existing in the taxonomy database
Please confirm and enter the name is ranked as “species” or other lower taxon (subspecies, variety, strain, etc.) and classified as “scientific name” (not svnonym, equivalent name or others).
When you have any objections against the description status of the taxonomy database, see here.
In cases of the organism names not found in the taxonomy database
To submit nucleotide sequence data to DDBJ or to search taxonomy database, please enter the organism name without any misspells.
- In case of unidentified species names
- Validly identified the scientific name of species
- Artificially constructed sequences
General rule to describe organism names
In general, an organism should be called its scientific name of species, however, when the species is not identified or not defined, it would be some tentative name instead of the scientific name of the species.
So, do NOT inappropriately select any organism name existing in taxonomy database.
Only when you can identify organism name from which your sequence has been obtained with no doubt AND
the organism name has been already registered to taxonomy database,
you can select the organism name existing in taxonomy database.
In general, sequence similarity of marker gene is not the only-one nor the best method to classify organisms and to identify species.
When you classify and identify species, you should consider comprehensive perspectives;
morphology, anatomy, physiology, behavior, molecular biology, culture condition, collection site, morphology, biochemical assessment and so on.
It is totally responsibility of its submitter for the contents of sequence data.
- The name of “unidentified organism” or “novel species” should be described with a tentative one
- They are totally different things, “Sequences of a gene are identical” and “Samples are derived from the same species”
- Sequence similarity of a marker gene is NOT absolute benchmark in phylogenetic relationship.
- The current standard for prokaryotic species identification is ANI (Average Nucleotide Identity) or dDDH (digital DNA-DNA hybridization) by using whole genome sequences
If the organism name that you submit is not in the database,
the name should be newly registered to the taxonomy database through DDBJ.
Such new organism names will be open to the public on taxonomy database
after the release of corresponding sequence data from DDBJ.
Before the release of corresponding sequence data from DDBJ, the organism name can not be available on taxonomy database.
Please do not hesitate to contact us when you like to update the organism name of your sequence data, once submitted to taxonomy database.
See also the page, Data Updates/Correction: after getting your accession number, when you like to update your data.
In principle, the organism name is required to be one of “scientific name” in taxonomy database.
When you have any objections against the description status of the taxonomy database, see here.
“Category” of organism name
During submission through Nucleotide Sequence Submission System you have to select “Category” of organism name.
Here shows a flowchart for judging “Category” of organism on Nucleotide Sequence Submission System
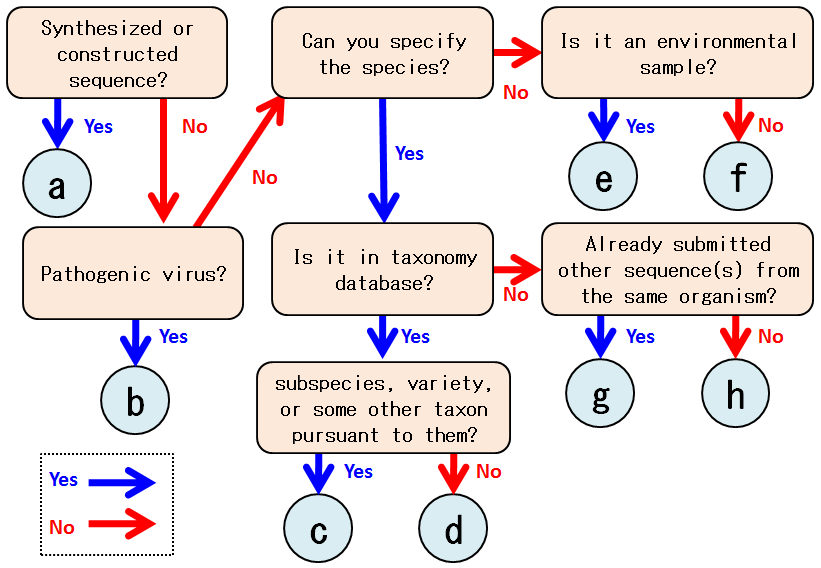
a. artificial constructed or synthesized sequence
Select “Artificially constructed or synthesized sequence” in the menu of [Category].
After referring to 4. Artificially constructed sequence, enter the name in the value of [Scientific name], appropriately.
b. virus
After finding the organism name from which your sequence was obtained, please select the name.
Please include the values of corresponding qualifiers,
/isolate (or /strain),
/serotype,
/collection_date,
/geo_loc_name,
/host etc., respectively.
See also the virus section.
In cases of pathogenic viruses, select “Pathogenic viruses” in the menu box of [Category].
c. trinominal name
Select “Valid subspecies or variety not registered in taxonomy database” in the menu box of [Category].
After referring to trinominal name, please include subspecies, variety, or strain etc. in the value of [Scientific name].
d. found in taxonomy database
After finding the organism name from which your sequence was obtained, please select the name.
You do not have to select any of [Category].
e. direct molecular isolation from a bulk environmental DNA sample
Select “Obtained by direct molecular isolation from environmental sample” in the menu box of [Category].
After referring to 3. Environmental samples, enter an organism name in the [Scientific name] box, appropriately.
Please include the values of corresponding qualifiers,
/isolation_source,
/isolate (or /clone,
/collection_date,
/geo_loc_name,
/host etc., respectively.
f. unidentified organism or novel species in proposing state
In case of a tentative name for an unidentified organism, select “Species is not identified”.
In case of a novel species in proposing state, select “Species is not identified” in the menu box of [Category].
After referring to 2. In case of unidentified species names, enter an organism name in the [Scientific name] box, appropriately.
g. not found in taxonomy database, but already submitted other sequence data of the organism
Select “The name is valid but not registered in taxonomy database” in the menu box of [Category].
Please input the same organism name as previously submitted (scientific name or tentative name) in the meantime before the valid publication of the species.
h. not found in taxonomy database, but validly identified the scientific name of species
Select “The name is valid but not registered in taxonomy database” in the menu box of [Category].
After referring to 1. For identified species, enter an organism name in the [Scientific name] box, appropriately.
See In cases of valid scientific names but not reported with any sequence data
Details to describe organism names
Though there are still some exceptions, followings are how to describe organism names for DDBJ submission.
If the application of the organism name to taxonomy database is required, during your sequence submission,
please let us know reference information.
1. For identified species
In principle, “organism name” is required to be a binomial name, i.e. the genus name and the species epithet, from which the sequence is obtained.
The species name should be described following international code of nomenclatures,
such as International Code of Zoological Nomenclature (ICZN),
International Code of Botanical Nomenclature (ICBN),
International Code of Nomenclature of Bacteria.
Example
Homo sapienstrinomial name
In cases of using trinomial name or the like, the name of subspecies, variety or serotype should be included in the organism name, if necessary.
Examples
Pan troglodytes troglodytes
Zea mays subsp. mays
Brassica oleracea var. alboglabra
Oryza sativa Japonica GroupAlso, the qualifier corresponding to /serotype or /strain is required for the source feature.
Example
/organism="Pan troglodytes troglodytes"
microbial genome
Previously, a strain name or some other lower taxon was required for the organism name of whole genomic scale sequences from microorganisms.
However, currently, the value of organism qualifier should be just a scientific name, in principle, even though for microbial genomes.
Describe a strain name in the /strain qualifier.
Example
/organism="Candida albicans"
/strain="WO-1"
cyanobacteria
In the cases of cyanobacteria, strains or some identifiers are included in the organism names, because of the specific research background of them.
Example
/organism="Geitlerinema earlei GSP174-1"
/strain="GSP174-1"
virus
For viruses, basically, we also accept scientific names, following the International Committee on Taxonomy of Viruses.
See also the virus section on Identifiers.
By 2017, we used informal names for frequently submitted pathogenic viruses including their strain names and serotypes in the description of organism name.
However, the rule has been discontinued for the future submissions.
Example
- Influenza A virus
The qualifiers corresponding to /collection_date, /geo_loc_name, /host, /isolate (or /strain) and /serotype are required for the source feature.
Please describe them with appropriate qualifiers as below example.
/collection_date="2007-11-01"
/geo_loc_name="Japan:Tokyo"
/host="Gallus gallus"
/isolate="A/chicken/Tokyo/2007"
/mol_type="viral cRNA"
/organism="Influenza A virus"
/serotype="H7N7"
hybrid
In cases of hybrids, the scientific names would be like as follows;
Examples
Rosa alba x Rosa corymbifera
Malus x domestica
Lilium hybrid division INot found in taxonomy database
In cases of valid scientific names but not reported with any sequence data
In principle, you can submit nucleotide sequence data with the organism name.
Please enter the organism name without any misspells.
If the name of genus or other upper taxa is not available on taxonomy database,
please tell us the lineage information of the organism.
As far as possible , please tell us the valid publication of the species.
Please note, even though the organism name has already been registered into the taxonomy database, when the data with the organism name have not yet been open to the public, you can not retrieve the name in the taxonomy database.
Notice) During submission via Nucleotide Sequence Submission System, please select “Not found in taxonomy database, but already registered in other sequence data “ for “Category” of organism name to indicate corresponding to the case.
Please tell us any of following items during your sequence submission.
Useful items for application of organism names to taxonomy database
Any objections against the description status of the taxonomy database
If you find any mistakes in the taxonomy database, please contact us to correct it.
In case of misspelling of the organism name, it will be easily corrected.
If you are preparing to intend reclassification of a series of organisms, so-called “combinatio nova”, the names on the taxonomy database can not be corrected before publishing your paper to claim the new classiffication.
So, in the meantime, please use the current names in the taxonomy database.
After published your paper, contact us to change/correct the scientific name. Then, we will check it again.
When you have any questions, objections, or comments for the classification of “scientific name”, “synonyms”, lineages, etc. on the taxonomy database,
DDBJ can ask managers of taxonomy database any modification based on evidence papers or references from you.
So, as far as possible, please provide the citation information that support your opinions and your comment on how it should be, because it often be highly technical content.
Please note, regarding issues such as interpretating phylogenetic lineages, the taxonomy database has a policy of adopting one of the theories to ensure the uniqueness of the data.
So, we may not be able to meet your request.
See also the description of the taxonomy database in detail.
Please tell us any of following items during your sequence submission.
Useful items for application of organism names to taxonomy database
2. In case of unidentified species names
2-1. In case of species not to be identified
If the scientific name is unclear and/or unidentified, we adopt a tentative name for the organism.
The tentative name is made up with the lineage (in many cases, genus names) that as far as submitters could specify.
Though, tentative names have been assigned to strains, redundant assignments are now avoided.
For prokaryotes
Format
- <genus name> sp. # prokaryotes
- <family (or upper) name> bacterium
- <family (or upper) name> archaeon
Examples
Acetobacter sp.
Acetobacteraceae bacterium
Methanomicrobiales archaeonFor eukaryotes
Format
- <genus (or upper) name> sp. # eukaryotes
Example
Aspergillaceae sp.
The qualifier corresponding to identifier, such as
/strain,
/isolate etc., is required for the source feature.
Though, tentative names have been assigned to strains, isolates, or other sample IDs, redundant assignments are now avoided.
/organism="Acetobacter sp."
/strain="ITDI2.1"In cases of whole genomic scale sequences from unicellular microorganisms,
some identifier, strain name or some other lower taxon, is required to include in the organism name.
The tentative name is made up with the lineage (in many cases, genus names) that as far as submitters could specify
and the lower taxon (in many cases, strain names).
Even included in the organism name, you should describe the identifier in the
/strain or some other corresponding qualifier.
/organism="Euglena sp. CR123"
/strain="CR123"Please use the same tentative name, when you submit any other data derived from the same organism.
2-2. In case of proposing a novel species
In cases of viruses, you can use the novel species name, as is, without the valid publication of the species that is named after The International Code of Virus Classification and Nomenclature (ICVCN).
However, in case of the virus named based on recent binominal ones and before valid publication, please use a tentative name in the following format.
Format
- <genus name> sp. ‘<candidate of species epithet>’
For other than viruses, no name confusable with the formal scientific name is acceptable before the valid publication.
In principle, during the stage proposing a novel scientific name, a tentative name like as unidentified cases is required
to keep unique correspondence between the organism and sequence data until established a new scientific name.
If you are to propose a novel species, in the mean time, we adopt an unique tentative name for the organism.
The tentative name is made up with the lineage (in many cases, genus names)
that as far as submitters could specify and the identifier (in many cases, strain names).
In cases of prokaryotes, a tentative name should be assigned to every strain.
Format
- <genus name> sp. <strain name> # prokaryotes
- <family (or upper) name> bacterium <strain name>
- <family (or upper) name> archaeon <strain name>
Examples
Acetobacter sp. ITDI2.1
Acetobacteraceae bacterium ITDI2.1In cases of eukaryotes, a tentative name should be assigned to every species candidate (operational taxonomic unit, OTU).
Though, tentative names have been assigned to strains, isolates, or other sample IDs, redundant assignments are now avoided.
Format
- <genus (or upper) name> sp. <OTU ID> # eukaryotes
Example
Oscinella sp. 1-AB-2020
Because we have to avoid some confusions, for example, two different organisms are mixed up.
Therefore, if you have any other sequence data derived from the same species candidate,
please use the same tentative name in the meantime before the valid publication of the species.
This is only a procedure to manage the submitted data to us, so, it is a different issue from any names that you use in the preparing paper and so on.
The tentative name in the taxonomy database is to be changed to the formal scientific name, after the valid publication of the species.
When the valid pulication, i.e. the paper to report a novel species, is published, please contact us to update the paper information (reference), the organism name and so on, and to release your sequence data.
See Data Updates/Corrections after Receiving Accession Number for your update request.
Also, the qualifiers corresponding to the identifier (such as /strain etc.) are required for the source feature to submit your sequence.
/organism="Acetobacter sp. ITDI2.1"
/strain="ITDI2.1"Please tell us any of following items during your sequence submission.
Useful items for application of organism names to taxonomy database
- taxonomic lineage
- proposing name for novel species
- already issued accession number
- process of sampling and/or sequencing
3. Environmental samples
Environmental samples are sequences derived by direct molecular isolation from a bulk environmental DNA sample
(by PCR, DGGE, or other anonymous methods) with no reliable identification of the source organism.
Though frequently confused, the term, ‘environmental samples’, does not mean “wild type”.
Please refer to environmental samples in detail.
Mixed culture derived from an environmental sample is also processed as a kind of environmental samples.
3-1 Environmental profile
For environmental sample, the organism name should be the lineage that as far as you can specify with the header “uncultured”.
Format
- uncultured bacterium # prokaryotes
- uncultured <genus name> sp. # prokaryotes
- uncultured <family (or upper) name> bacterium # prokaryotes
- uncultured <genus (or upper) name> # eukaryotes
Examples
uncultured bacterium
uncultured Acetobacter sp.
uncultured alpha proteobacterium
uncultured Bacillaceae bacterium
uncultured Aspergillus
uncultured AspergillaceaeIn cases of environmental samples, the qualifier,
/environmental_sample, is required for source feature.
Also, /isolation_source and some other qualifiers should be used to describe the process and conditions of sample isolation.
The qualifiers, /collection_date,
/geo_loc_name and
/host
are also useful.
For the identifier, use /isolate qualifier.
When you cloned the sample sequence, use /clone qualifier, instead of /isolate qualifier.
/clone="4-11"
/collection_date="2007"
/environmental_sample
/geo_loc_name="Japan: Shizuoka"
/isolation_source="PCR-derived sequence from sediment"
/mol_type="genomic DNA"
/organism="uncultured Acetobacter sp."If the name is not available on taxonomy database (TXSearch), please tell us any of following items during your sequence submission.
Useful items for application of organism names to taxonomy database
3-2 Metagenome Assembly
: When you submit sequences of Metagenome-Assembled Genome (MAG), some single taxa can be assigned by the software tools, so use the names of species or lower ranks (ex. Agrobacterium tumefaciens) for the /organism qualifiers.
If the scientific name of MAG is unclear and/or unidentified, we adopt a tentative name for the organism.
The tentative name is made up with the lineage (in many cases, genus names) that as far as submitters could specify.
Format
- <genus name> sp. # prokaryotes
- <family (or upper) name> bacterium
- <family (or upper) name> archaeon
- <genus (or upper) name> sp. # eukaryotes
Examples
Agrobacterium sp.
Rhizobiaceae bacterium
Methanomicrobiales archaeon
Aspergillaceae sp.In case of MAG, the qualifiers,
/environmental_sample,
/isolation_source, and
/isolate (as an identifier),
are required for source feature like as other environmental samples.
Additionally, the /metagenome_source qualifier is also required for source feature of MAG.
For /metagenome_source qualifier, please input some of operative
metagenome taxonomy nodes reflecting the resource of the environmental sample, e.g. “soil metagenome” .
See NCBI site about usages of metagenome names.
Useful items for application of organism names to taxonomy database
4. Artificially constructed sequences
In general, artificially constructed sequences are uniformly named “synthetic construct” or “eukaryotic synthetic construct”.
Sometimes, vector names or something like that are described ‘as is’ in the organism name.
When you like to submit sequences of only some specific genes, use “synthetic construct” or “eukaryotic synthetic construct”.
If you like to submit complete sequences of vectors or the likes, please give some name to them.
To distiguish with natural plasmids. do not use the word, “plasmid “.
Examples
Cloning vector pAP3neo
Expression vector pAMPIf the name is not available on taxonomy database (TXSearch), please tell us any of following items during your sequence submission.
Useful items for application of organism names to taxonomy database
Useful items for application of organism names to taxonomy database
If the organism name that you submit with your sequence data is not in the database, it must be newly registered to the taxonomy database through DDBJ.
taxonomic lineage
Please tell us the taxonomic lineage of the organism from which your sequence has been obtained, as far as inferable.
valid publication for species
If the scientific name of species has been publicized in papers,
please tell us the references for the species.
proposing name for novel species
If it is really novel species and not yet published, please tell us proposing name for the novel species
for the tracking purpose at taxonomy database. In addition, please send E-mail to DDBJ update,
 ,
when it is published or particularly if the name changes.
,
when it is published or particularly if the name changes.
already issued accession number
If you have already submitted other sequence data to INSDC (DDBJ/ENA/GenBank or BioSample)
derived from the same organism of present submission and
the previous data has not yet been published,
please tell us the accession number(s) of your previous data.
process of sampling and/or sequencing
It would be helpful if you tell us some information such as process of sampling, sequencing and so on.
expected usage
In cases of artificial constructed sequences, please tell us how to use them.