DDBJ Annotated/Assembled Sequences
Data Submission from Genome Project
This page shows steps of genome sequencing, categories of sequence data and their correspondences, briefly.
The schematic diagram below indicates phases of typical genome sequencing strategies.
Also, please submit to
BioProject and
BioSample, in case of large scale genome sequencing project.
Important: Data submission of human subjects research
See also the following sites.
- Finished level genomic sequences
- Haplotype
- INSDC standards for genome assembly submission
- Metagenome Assembly and MAG
- Single amplified genome SAG
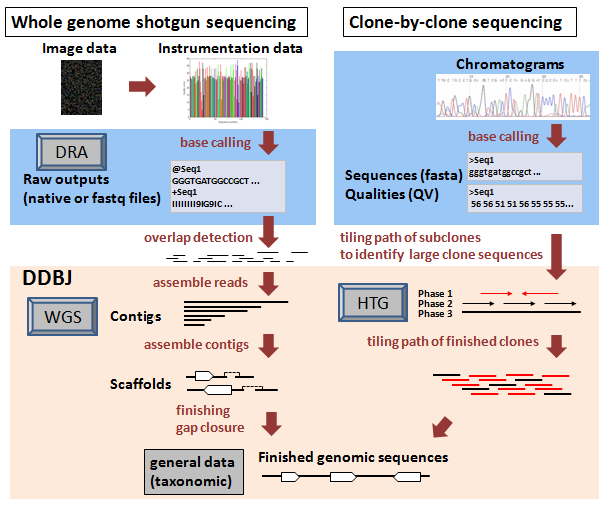
- [DRA] Raw outputs: data generated by next-generation sequencers
- In case of output data generated by next-generation sequencing platforms, submit to DDBJ Sequence Read Archive (DRA).
- [WGS] Contigs: assemblies (overlapping reads)
- In case of assemblies (i.e. overlapping reads) that are appropriately assembled sequences excluded redundancy from raw reads, submit to Mass Submission System as WGS data.
- [HTG] draft sequences of large clones
- In case of unfinished level, draft sequences of BAC, YAC or fosmid clones, submit to Mass Submission System as HTG data.
- Finished level genomic sequences
- Submit to Mass Submission System as general data or complete genomes.