DDBJ Annotated/Assembled Sequences
INSDC standards for genome assembly submission
The original site; INSDC standards for genome assembly submission 2013.08.14 version
Genome assemblies comprise a number of possible layers of information, including reads, contigs, scaffolds and chromosomes (see figure I). This document lays out the requirements for submission of genome assembly information into INSDC databases.
Figure I. Three typical assembly processes
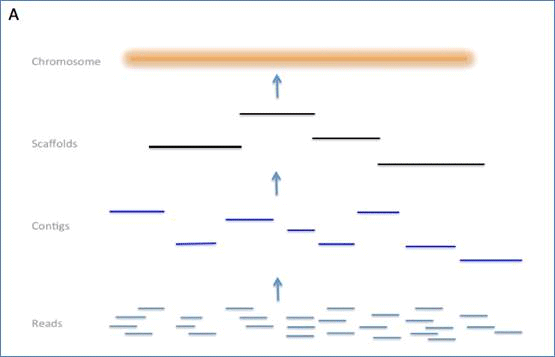
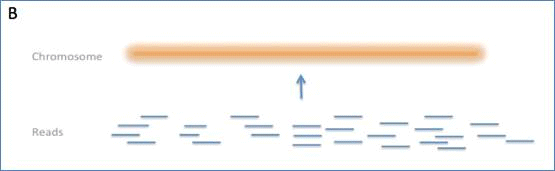
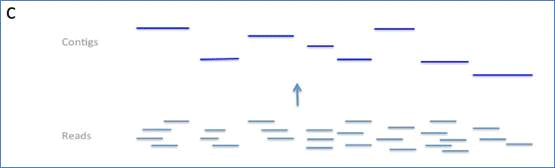
Figure I. The figure shows three typical assembly processes and the
layers of information that they yield.
A) Clone-based assembly with scaffolding and finishing steps.
B) Shotgun assembly direct to chromosomes.
C) Partial assembly to contigs only.
Table I. New genome assembly submissions
| Component | Level | Comment |
| Reads | Recommended | Complete read and quality data |
| Read to contig mapping | One of, as appropriate, optional | e.g. BAM alignment of reads to contigs |
| Read to chromosome mapping | e.g. BAM alignment of reads to new chromosome | |
| Contigs | At least one layer mandatory | |
| Scaffolds | ||
| Chromosomes | ||
| Scaffold to chromosome mapping | Mandatory if both layers are present | e.g. AGP file |
| Contig to scaffold mapping | Mandatory if both layers are present | e.g. AGP file |
| Assembly description | Mandatory | Brief information relating to assembly and future plans |
| Functional annotation | Optional |
Consistent with the variety of assembly processes, submitters to INSDC approach with data for the layers in different combinations of layers. Tables I and II shows requirements for new genome assembly submissions and updates to existing assembly submissions.
Table II. Update to existing genome assembly
| Component | Level | Comment |
| Reads | Recommended | Complete read and quality data |
| Read to contig mapping | One of, as appropriate, optional | e.g. BAM alignment of reads to contigs |
| Read to chromosome mapping | e.g. BAM alignment of reads to new chromosome | |
| Contigs | At least one layer mandatory, with highest layer no lower than for existing assembly | |
| Scaffolds | ||
| Chromosomes | ||
| Scaffold to chromosome mapping | Mandatory if both layers are present | e.g. AGP file |
| Contig to scaffold mapping | Mandatory if both layers are present | e.g. AGP file |
| Assembly description | Mandatory | Brief information relating to assembly and future plans |
| Regenerated (or lifted-over) functional annotation | Recommended | If associated with existing assembly |
| Coding annotation mappings between old and new assemblies | Recommended where functional annotation is provided for the updated assembly | Typically through INSDC protein ID mappings |
Third party genome assembly submissions and updates, in which the submitting group does not hold complete ownership of data, are subject to existing third party annotation rules, including the requirement for presentation of the new/updated genome assembly in a peer reviewed publication prior to public release from INSDC.