TXSearch
TXSearch is a NCBI Taxonomy retrieval system provided by DDBJ. When you submit nucleotide sequence data to International Nucleotide Sequence Database(INSD), you need to use the organism name description based on NCBI Taxonomy.
Search (from the web form)
Top page (Tree screen):http://ddbj.nig.ac.jp/tx_search/
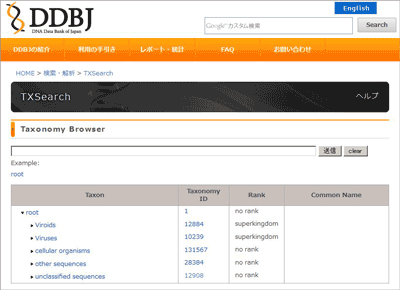
Search from the Web Form
Enter the words and/or numericals into the box, then click the “send” button or press the “Enter” key.
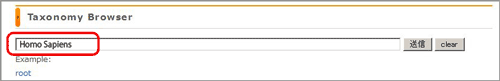
-
“Wild Card” (“?” stands for any single character, and “*” for any number of characters) is available.
-
Boolean Operators (AND, OR, NOT) can be entered as follows.
AND “space” or “AND” or “&&” OR “OR” or “ ” NOT “NOT” -
To use the “Example” is an easy way.
- (1) Click the “Example” under the search box, some examples are displayed.
- (2) Select the one and click it, then its text is entered into the search box.
- (3) Modify the text to suit your purpose. (e.g. organism names, taxonomy_id)
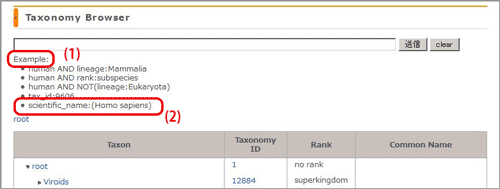
-
Default search fields are as follows.
tax_id anamorph in_part scientific_name teleomorph includes common_name authority misnomer synonym blast_name misspelling acronym equivalent_name type_material If you search some particular fields, specify the field by refine search.
-
Because the maximum number of results displayed in the screen is fixed in 1000 (in ascending order of scientific_name), to view the all results, please execute the refine search to narrow the number down to 1000.
Refine Search
You can narrow the scope of your search by specifying some particular fields, and/or combining some field searches.
- When the result is more than 1000, please execute the refine search so that less than 1000 is included in the search result (In this system the maximum result display number is fixed as 1000 which is unmodifiable)
- The description to search a specific field is, field name:value (e.g. tax_id:9606)
- Multiple fields
Connect “field name:value” units, separating by space (e.g. scientific_name:(Homo Sapiens) && rank:species) -
Following fields can be specified
Searchable fields tax_id
scientific_name
common_name
synonym
acronym
anamorph
teleomorph
authority
blast_name
equivalent_name
in_part
includes
misnomer
misspelling
type_materialWhen the field name is specified, specified field is searched.
When the field is not specified, all these field is searched.lineagerank To search these fields, you need to specify the field-name and its value. -
Exact matching, please specify the field name as follows.
scientific_name_ex common_name_ex synonym_ex acronym_ex anamorph_ex teleomorph_ex authority_ex blast_name_ex equivalent_name_ex in_part_ex includes_ex misnomer_ex misspelling_ex type_material_ex
Search Examples
| Text | what you search |
|---|---|
| human AND lineage:Mammalia | search the taxon which contains “human” in the default search fields, and linage is “Mammalia” |
| human AND rank:subspecies | search the taxon which contains “human” in the default search fields, and rank is “subspecies” |
| human AND NOT(lineage:Eukaryota) | search the taxon which contains “human” in the default search fields, and linage is NOT “Eukaryota” |
| tax_id:9606 | tax_id is 9606 |
| Homo Sapiens | search the taxon which contains both “Homo” AND “Sapiens” in the default fields |
| scientific_name:”Homo sapiens” | search the taxon whose scientific_name includes “Homo sapiens” |
| scientific_name:(Homo sapiens) | search the taxon which contains both “Homo” and “sapiens” in the scientific_name field. |
| scientific_name:Homo sapiens | search the taxon which contains both “Homo” in the scientific_name field, and contains “sapiens” in the default search fields. |
| scientific_name_ex:”Homo sapiens” | search the taxon whose scientific_name is “Homo sapiens” (Exact search). (Note) When the search words contains a space, the words should be enclosed in double quotation marks. |
| 50000 | search the taxon which contains “50000” in the default search fields. (In this case, both the taxon which contains “50000” in the name, such as “Crinipellis sp. GDGM 50000”, and “Sarracenia alabamensis” whose tax_id is “50000” are retrieved. To search either one, you should specify the field name you want to retrieve.) |
| tax_id:(741158 OR 63221) or tax_id:(741158 || 63221) |
searchs tax_id field either 741158 OR 63221 |
| scientific_name:(Acetobacter c*) | search the taxon which contains both “Acetobacter” and “the strings” containing any number of characters starting with “c” in the scientific_name field. |
| scientific_name:(Acetobacter sp*) | search the taxon which contains both “Acetobacter” and “the strings” containing any number of characters starting with “sp” in the scientific_name field. |
| tax_id:960? | search the tax_id field in 960? (? is one character) |
Search Result
Search Result (Taxon Information screen)
This is the result screen of the search by the web form.
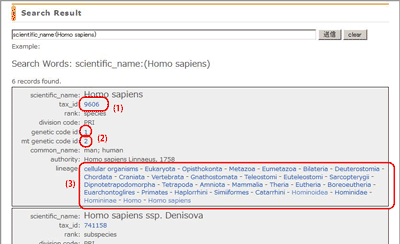
- Scientific name, taxonomy id, rank, division code, genetic code, mitochondrial genetic code, names other than scientific name, and lineage of specified taxonomy id’s taxon are displayed.
- taxonomy id links to NCBI Taxonomy Browser.
- genetic code id, mitochondrial genetic code id links to Genetic code table.
- Each name in the lineage line links to the tree.
Tree Screen
Click the name in the lineage, then the tree will be displayed on the screen.
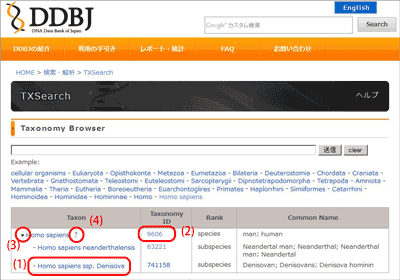
- (1) scientific name links to Taxon Information screen.
- (2) taxonomy id links to NCBI Taxonomy Browser.
- (3) Delta icon left side of scientific name developes taxonomic tree of lower rank.
- (4) The right hand arrow icon of top
scientific name developes upper level tree.
If you click it, more 1 ranks are displayed. - You can change the number of ranks displayed on the browser by the modification of depth parameter value in the URL.
Method including taxonomy_id into the URL
You can make the URL which include taxonomy_id. Using this, you can display the search result directly, and/or link to the Taxon Information page.
When including taxonomy_id in URL
| Parameters | Details | Examples |
|---|---|---|
| id (required item) | specifies target taxonomy id. |
|
| view | View of Tree of Taxon information. In case of not specified, automatically tree view is selected. |
|
| depth | specifies number of hierarchy. Default is 2. |
|