VecScreen
About VecScreen
VecScreen is a tool that can detect foreign DNAs such as vector, linker, adapter, and primer regions (we call vector contamination in this document) involved in nucleotide sequences (query) by using blast search against vector sequence database. The main unit of this tool is vecscreen program obtainable from NCBI.
Available Internet browsers
We confirmed that IE8 or newer, latest version of Firefox, Chrome, and Safari can work correctly. If you use VecScreen by an older version of Internet browser, you may meet some trouble for executing the search.
How to use
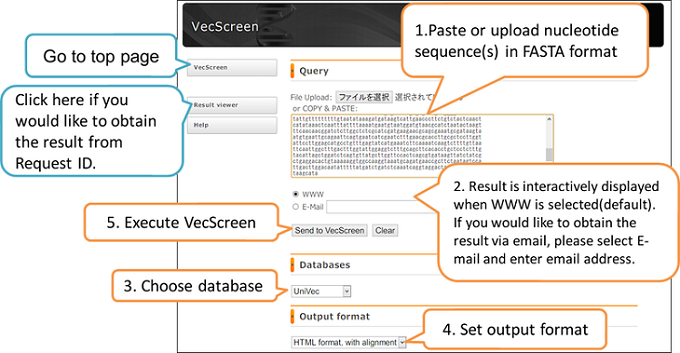
Query
- Please enter the query sequence(s) in FASTA format. Please put query sequence(s) on the text box or upload the query sequence(s) through “File Upload”.
- When query consist of multiple sequences, you need to add each sequence name by using “>” (right angle bracket) to the line just before each sequence (multi FASTA format, see example below).
- When you investigate a single sequence, you do not have to add a name to the sequence, however, if you would like to assign a name to the query, you should describe the name with “>” (right angle bracket) before the first line of the sequence.
FASTA format
>my query sequence 1
caccctctcttcactggaaaggacaccatgagcacggaaagcatgatccaggacgtggaa
gctggccgaggaggcgctccccaggaagacagcagggccccagggctccaggcggtgctg
gttcctcagcctcttctccttcctgctcgtggcaggcgccgccac
Multiple sequences (multi FASTA)
>my query sequence 1
caccctctcttcactggaaaggacaccatgagcacggaaagcatgatccaggacgtggaa
gctggccgaggaggcgctccccaggaagacagcagggccccagggctccaggcggtgctg
gttcctcagcctcttctccttcctgctcgtggcaggcgccgccac
>my query sequence 2
ggccagggcacccagtctgagaacagctgcacccgcttcccaggcaacctgcctcacatg
cttcgagacctccgagatgccttcagcagagtgaagactttctttcaaatgaaggatcag
ctggacaacatattgttaaaggagtccttgctggaggactttaag
>my query sequence 3
atgggtctcacctcccaactgcttccccctctgttcttcctgctagcatgtgccggcaac
tttgcccacggacacaactgccatatcgccttacgggagatcatcgaaactctgaacagc
ctcacagagcagaagactctgtgcaccaagttgaccataacggac
When the query sequence size is too big (a large number of sequences, or each sequence is very long), the result might not be displayed on the web screen normally because of the following reasons.
- Internet browser cannot display the result because of vastness of result size.
- VecScreen terminate abnormally because of lacks of memory.
- Timeout occurs because of a long time for analysis.
In such a case, you should reduce number of nucleotide sequences or shorten the sequence length and retry the search.
WWW or Email
When “WWW” is selected, the result is interactively displayed on the screen. If you would like to obtain the result by an email, please select “E-mail” and enter your email address correctly. WWW is selected by default.
Databases
Please select a vector database either UniVec or UniVec_Core. UniVec is selected in the default. The system uses the same database that can be obtained from https://ftp.ncbi.nih.gov/pub/UniVec/.
- UniVec
- UniVec is a non-redundant vector database consists of many vector, linker, adapter, and primer sequences. When UniVec is selected, users must remove false positive hit from the result by their manual check. You should choose UniVec when you would like to obtain maximum detection of vector contamination by allowing false positive hit.
- UniVec_Core
- UniVec_Core is a subset of UniVec database. UniVec_Core is designed so that the users can automatically detect vector contamination without manually reviewing the result. It is expected that the result involves minimum size of false positive hits.
UniVec_Core contains oligonucleotide and vector sequences from bacteria, phage, yeast, and synthetic construct and does not involve vector sequences such as mammalian-derived sequence. Therefore, UniVec_Core is sometimes unable to detect vector contamination that UniVec can do it.
Output format
Please select a type of result format. “HTML format, with alignment” is set in the default.
- HTML format, with alignment
- Result is obtained in HTML format. Graphic viewer showing vector contamination region on query, summary of the result, and alignment are displayed((example).
- HTML format, no alignment
- Result is obtained in HTML format. Graphic viewer showing vector contamination region on query and summary of the result are displayed(example).
- Text list, with alignment
- Result is obtained in TEXT format. Summary of the result and alignment are displayed(example).
- Text list, no alignment
- Result is obtained in TEXT format. Summary of the result is only displayed(example).
There is no parameter settings
VecScreen does not have option for blastn parameters. The tool executes
vecscreeen program that is obtained from NCBI. For details, please refer
the links below.
//www.ncbi.nlm.nih.gov/tools/vecscreen/about/
//www.ncbi.nlm.nih.gov/tools/vecscreen/univec/
Execution (Send to VecScreen)
Vector contamination check starts when you click “Send to vecscreen”. When you click “Clear” query sequence(s) is erased.
Result screen
According the scores and e-value, the result are displayed with the classification of three categories, Strong match, Moderate match, and Weak match, which indicates the order of the probability of vector contamination (Strong > Moderate > Weak). The <50-bp region between the two vector hit position or the region from vector hit position to the end are also detected as Suspect Origin which suggests possible vector contamination. For details, please refer to the URL below. //www.ncbi.nlm.nih.gov/tools/vecscreen/about/#Categories
Determination of the position of vector contamination
If you would like to carry out the detection of vector contamination automatically, the regions that are indicated by Strong match and Moderate match are usually taken as vector contamination. Weak match shows a possibility of vector contamination and Suspect origin also suggests the contamination. The two categories, Weak match and Suspect origin doesn’t always show the vector contamination.
Strictly speaking, you should determine the correct position of the vector contamination from the VecScreen results by taking into consideration of vector, adapter, linker, primer sequences used by cloning experiment and cloning method.
Please refer to the URL below for detailed explanation of the result. //www.ncbi.nlm.nih.gov/tools/vecscreen/interpretation/
Notes when the result is not correct
VecScreen is not a perfect tool that can detect entire vector contamination. When query sequences that are not covered with UniVec database were used, it is expected that the VecScreen cannot detect at all. Or VecScreen may underestimate vector contamination region or predict the results as lower categories even though VecScreen can detect a similar sequences from UniVec. You should carry out vector trimming with consideration of foreign DNA and method that are actually used for the cloning experiment.
Exceptions
UniVecデータベースの構成上,以下の配列をクエリーに用いた場合もベクターコンタミとして検出されることになりますのでご承知おきください。 詳しくは,//www.ncbi.nlm.nih.gov/tools/vecscreen/interpretation/#Exceptions をご覧ください。
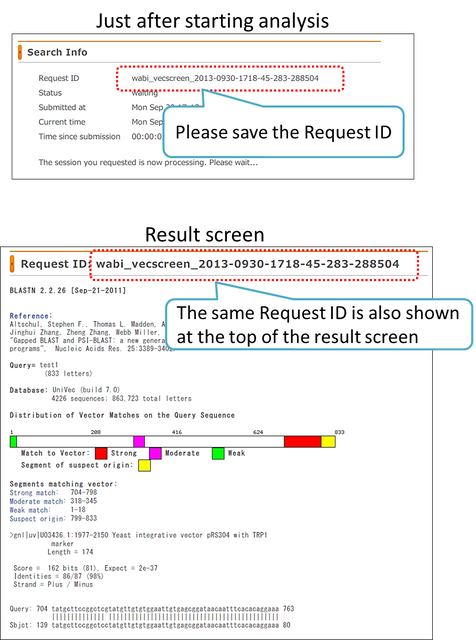
Request ID
After pressing the “Send to VecScreen” button, Request ID is displayed on the screen. The same ID is also shown at the top of the result screen. Do not forget the ID because it is necessary for using the “Result Viewer” and/or inquiring to DDBJ for your search. You can view your result using “Request ID” at any time until the cutoff date even if the browser is closed.
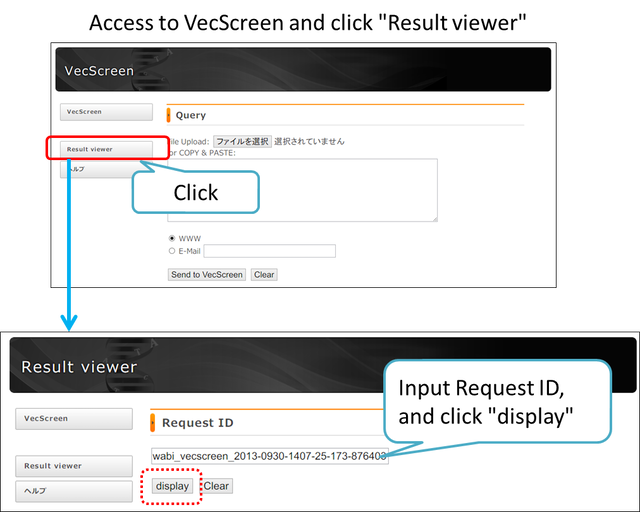
Result viewer
You can obtain the result from Request ID. Please click “Result viewer” on the left side of VecScreen to change the screen. Then, enter the Request ID and click “display”.
Time limit for viewing a result
The result is eliminated in 7 days after execution.