getentry
About getentry
- getentry is the DDBJ flat file search system by accession numbers.
- getentry is, as well as a simple web form, available by WebAPI program which calls up the data directly.
- WebAPI also provides a gethistory (revision history search) function.
- Keyword search of DDBJ flat file is now avialable in ARSA
- DRA data retrieval is unavilable in getentry, please refer DDBJ Search
Search from Web Browser
URL : https://getentry.ddbj.nig.ac.jp/top-e.html
Default value
| ID | Accession Number |
|---|---|
| Database | DNA database:DDBJ/EMBL/GenBank INSD. |
| Output format | flatfile(DDBJ) |
| Result | html |
| Limit | 10 |
ID
Input the Accession Number(s). Multiple number search, range search, and version seach are available.
| version search |
|
AB669632.1 AB669632.2 |
|---|---|---|
| multiple number search |
|
AB669632.1,AB669632.2,AB669633.1,AB669633.2 AK377101 - AK377200,AK377210,AK377211 |
| range search |
|
FY782000-FY7830 AK377101 - AK377200,AK377211- AK388100 |
Database and Output format
Select the database. The output format should be selected from either “database specific format” or FASTA.
DNA database
Database
| DDBJ/EMBL/GenBank | International Nucleotide Sequence Databases(INSD) |
|---|---|
| MGA | Mass sequence for Genome Annotation(MGA) |
DDBJ/EMBL/GenBank includes the following databases.
- DDBJ periodical release
- daily updates
- TPA(Third Party Data)
TPA data are not contained in the DDBJ releases.
DDBJ terminated accepting new submission of MGA data.
Please refrer the Latest Release Information, for the current status of searchable databases and related information.
Output format
| flatfile(DDBJ) | DDBJ flat file format |
|---|---|
| Total nt seq FASTA | nucleotide total sequence in FASTA format |
| CDS Amino acid seq FASTA | amino acid translated sequence of CDS region in FASTA format |
| CDS nt seq FASTA | nucleotide sequence of CDS region in FASTA |
| INSD-XML_v1.4 | INSD-XML_v1.4 format |
These 5 format types are selectable only after specification of “DDBJ/EMBL/GenBank”.
When the MGA is selected as the target database, only “flatfile” can be specified.
Protein database
Database
| UniProt | Amino acid database (UniProt/Swiss-Prot and UniProt/TrEMBL) |
|---|---|
| PDB | 3D structure database of protein |
| DAD | translated sequences database extracting from DDBJ |
| Patent | Amino acid data from JPO and KIPO. |
Please refer the links in the above table for the current status of databases and related information.
Output Format
| default | Database specific format | ||
|---|---|---|---|
| FASTA | Amino acid seq FASTA | Amino acid sequence in FASTA format | available in UniProt, DAD, Patent |
| Nucleotide seq FASTA (for DAD) | Nucleotide sequence in FASTA format | DAD limited | |
| seqres | PDB specific amino acid sequence FASTA format | PDB limited | |
Output format of Protein database search differs depending on the selected target databases.
Filetype of the Result
| default | html |
|---|---|
| html | HTML (with link to ACCESSION, ORGANISM, etc) |
| text | text |
| gz | gzip compressed |
The name of the gzip files corresponding to the specified formats are as follows.
| [DNA]flatfile | flatfile.txt.gz |
| [DNA]xml | insd.xml.gz |
| [DNA]fasta | fasta_na.txt.gz |
| [DNA]trans | cds_aa.txt.gz |
| [DNA]cds | cds_na.txt.gz |
| [Protein]flatfile | flatfile.txt.gz |
| [Protein]fasta | fasta_aa.txt.gz |
| [Protein]cds | cds_aa.txt.gz |
Limit(upper limit of the data acquisition)
| default | 10 entries |
| specify the number | specified number of entries |
| 0? | no limit |
Search by WebAPI
getentry is available from WebAPI program, as well as a simple web form, which calls up the data directly.
Program
WebAPI of getentry consists of two following programs.
| getentry | Get the flat file of the specified accession number (ID of entries in the database) |
|---|---|
| gethistory | Get the revision history of the specified accession number (ID of entries in the database) Revision history of the amino acid sequence derived from the Patent Office has not been taken so far. |
How to Specify the Parameter
There are the following 2 methods.
| GET method | https://getentry.ddbj.nig.ac.jp/getentry?database=database name&accession_number=accession number&additional parameters (optional) |
| smart URL | https://getentry.ddbj.nig.ac.jp/getentry/database name/accession number https://getentry.ddbj.nig.ac.jp/getentry/database name/accession number/?additional parameters (optional) https://getentry.ddbj.nig.ac.jp/getentry/database name/accession number/revision ID/?additional parameters (optional) |
example
-
AB601234 by Get method
https://getentry.ddbj.nig.ac.jp/getentry?database=ddbj&accession_number=AB601234 -
BD500001(amino acid sequence data originated from Patent Office) by smart URL
https://getentry.ddbj.nig.ac.jp/getentry/patent_aa/BD500001
Specifiable Parameters in getentry
accession 番号(mandatory):Specify the accession number.
| version number |
|
|---|---|
| multiple accession search |
|
| range search |
|
example (upper:Get method / lower: smart URL)
-
AB055395
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=AB055395
https://getentry.ddbj.nig.ac.jp/getentry/na/AB055395 -
multiple accessions(multiple items, view 100 results)
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=AK377101 - AK377200,AK377210,AK377211-AK388100&limit=100
https://getentry.ddbj.nig.ac.jp/getentry/na/AK377101 - AK377200,AK377210,AK377211- AK388100?limit=100 -
multiple accessions(range search, view 1000 results)
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=FY782000-FY783000&limit=1000
https://getentry.ddbj.nig.ac.jp/getentry/na/FY782000-FYFY783000?limit=1000 -
multiple accessions with version numbers:
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=AB669632.1,AB669632.2,AB669633.1,AB669633.2,AB669634.1
https://getentry.ddbj.nig.ac.jp/getentry/na/AB669632.1,AB669632.2,AB669633.1,AB66963.2,AB669634.1 -
range search with version numbers:version search is ignored(latest version is searched) https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=AB669632.1-AB6696356.1
https://getentry.ddbj.nig.ac.jp/getentry/na/AB669632.1-AB6696356.1
database(optional):Specify the database for searching.
| DNA | na | DDBJ/EMBL/GenBank | International Nucleotide Sequence Databases(INSD) |
|---|---|---|---|
| mga | MGA | Mass sequence for Genome Annotation(MGA) | |
| Protein | aa | DAD, Patent, UniProt, PDB | search these 4 dbs in this order |
| uniprot | UniProt | Amino acid database (UniProt/Swiss-Prot and UniProt/TrEMBL) | |
| pdb | PDB | 3D structure database of protein | |
| dad | DAD | translated sequences database extracting from DDBJ | |
| patent_aa | Patent | Amino acid data from JPO and KIPO |
Omitting the database specification is processed as t
DDBJ/EMBL/GenBank includes the following databases.
- DDBJ periodical release
- DDBJ daily updates
- accession numbers consisting of “4 letters + 8 digits”
(WGS(Whole Genome Shotgun), some TSA(Transcriptome Shotgun Assembly)) - TPA(Third Party Data)
4 letters + 8 digits” accession numbers(WGS and TSA) and TPA are not contained in the DDBJ releases.
Please refrer Latest Release Information , for the current status of searchable databases and related information.
example (upper:Get method / lower: smart URL)
-
TSA( IAAA01000001) search
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=IAAA01000001
https://getentry.ddbj.nig.ac.jp/getentry/na/IAAA01000001 -
WGS(BAET01000001) search
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=BAET01000001
https://getentry.ddbj.nig.ac.jp/getentry/na/BAET01000001 -
AB628096, not selecting the database
https://getentry.ddbj.nig.ac.jp/getentry?accession_number=AB628096 -
MGA( AAAAA0000001)search
https://getentry.ddbj.nig.ac.jp/getentry?database=mga&accession_number=AAAAA0000001
https://getentry.ddbj.nig.ac.jp/getentry/mga/AAAAA0000001 -
Patent amino acid data search
https://getentry.ddbj.nig.ac.jp/getentry?database=patent_aa&accession_number=DI500001
https://getentry.ddbj.nig.ac.jp/getentry/patent_aa/DI500001
https://getentry.ddbj.nig.ac.jp/getentry?database=aa&accession_number=BD500001
https://getentry.ddbj.nig.ac.jp/getentry/aa/BD500001 -
DAD search
https://getentry.ddbj.nig.ac.jp/getentry?database=dad&accession_number=AB000714-1
https://getentry.ddbj.nig.ac.jp/getentry/dad/AB000714-1 -
UniProt search
https://getentry.ddbj.nig.ac.jp/getentry?database=aa&accession_number=P06213
https://getentry.ddbj.nig.ac.jp/getentry/aa/P06213
https://getentry.ddbj.nig.ac.jp/getentry?database=uniprot&accession_number=P06213
https://getentry.ddbj.nig.ac.jp/getentry/uniprot/P06213
revision(optional): Search the revised entry at the specified time.
| general | yyyy-MM-dd hh:mm:ss |
|---|---|
| release | yyyy-MM-dd hh:mm:ss release |
example (upper:Get method / lower: smart URL)
-
AB479935 as of 2011-05-31 23:07:30
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=AB479935&revision=2011-05-31 23:07:30
https://getentry.ddbj.nig.ac.jp/getentry/na/AB479935?revision=2011-05-31 23:07:30 -
Update history is avilable using gethistory.
https://getentry.ddbj.nig.ac.jp/gethistory?database=na&accession_number=AB479935
https://getentry.ddbj.nig.ac.jp/gethistory/na/AB479935
format(optional)
| default | flatfile |
|---|---|
| flatfile | DDBJ flat file format |
| xml | INSDSeq-XML version 1.4 |
| fasta | [DNA]Total nt seq FASTA |
| [Protein]Amino acid seq FASTA | |
| trans | [DNA]CDS amino acid seq FASTA |
| cds | [DNA] CDS nt seq FASTA |
| [DAD] Nucleotide seq FASTA (for DAD) | |
| seqres | [Protein] PDB amino acid |
Available output formats by specified database are as follows.
| DNA database | |
|---|---|
| DDBJ / EMBL / GenBank / MGA | flatfile(DDBJ), Total nt seq FASTA, CDS amino acid seq FASTA, CDS nt seq FASTA, INSD-XML_v1.4 |
| Protein database | |
|---|---|
| UniProt | default, Amino acid seq FASTA |
| PDB | default, seqres |
| DAD | default, Amino acid seq FASTA, nt seq FASTA |
| Patent | default, Amino acid seq FASTA |
example (upper:Get method / lower: smart URL)
- AB628096 in the flatfile format
https://getentry.ddbj.nig.ac.jp/getentry?accession_number=AB628096
https://getentry.ddbj.nig.ac.jp/getentry/na/AB628096
LOCUS AB628096 390 bp RNA linear VRL 24-FEB-2012
DEFINITION Human rhinovirus C gene for polyprotein, partial cds, strain:
HRV/Yamaguchi/2010/89.
ACCESSION AB628096
VERSION AB628096.1
KEYWORDS .
SOURCE Human rhinovirus C
ORGANISM Human rhinovirus C
Viruses; ssRNA positive-strand viruses, no DNA stage;
Picornavirales; Picornaviridae; Enterovirus.
REFERENCE 1 (bases 1 to 390)
AUTHORS Kobayashi,M., Arakawa,M., Okamoto,R., Tsukagoshi,H., Ryo,A.,
Mizuta,K., Hasegawa,S., Hirano,R., Wakiguchi,H., Kudo,K.,
Tanaka,R., Morita,Y., Noda,M., Kozawa,K., Ichiyama,T., Shirabe,K.
and Kimura,H.
TITLE Direct Submission
JOURNAL Submitted (06-MAY-2011) to the DDBJ/EMBL/GenBank databases.
Contact:Miho Kobayashi
Gunma Prefectural Institute of Public Health and Environmental
Sciences; Kamiokimachi 378, Maebashi, Gunma 371-0052, Japan
REFERENCE 2
AUTHORS Arakawa,M., Okamoto-Nakagawa,R., Toda,S., Tsukagoshi,H.,
Kobayashi,M., Ryo,A., Mizuta,K., Hasegawa,S., Hirano,R.,
Wakiguchi,H., Kudo,K., Tanaka,R., Morita,Y., Noda,M., Kozawa,K.,
Ichiyama,T., Shirabe,K. and Kimura,H.
TITLE Molecular epidemiological study of human rhinovirus species A, B
and C from patients with acute respiratory illnesses in Japan
JOURNAL J. Med. Microbiol. 61, 410-419 (2012)
REMARK DOI:10.1099/jmm.0.035006-0
COMMENT
FEATURES Location/Qualifiers
source 1..390
/collection_date="14-Jan-2010"
/country="Japan:Yamaguchi"
/db_xref="taxon:463676"
/host="Homo sapiens"
/isolation_source="Nasopharyngeal swab"
/mol_type="genomic RNA"
/organism="Human rhinovirus C"
/strain="HRV/Yamaguchi/2010/89"
CDS 1..>390
/codon_start=1
/product="polyprotein"
/protein_id="BAK22546.1"
/transl_table=1
/translation="MGAQVSKQNVGSHENSVSATGGSVIKYFNINYYKDSASSGLTKQ
DFSQDPSKFTQPLAEALTNPALMSPTVEACGMSDRLKQITIGNSTITTQDTLNSILAY
GEWPKYLSDLDASSVDKPTHPETSSDRF"
BASE COUNT 124 a 95 c 77 g 94 t
ORIGIN
1 atgggcgcac aggtgagcaa gcaaaatgtc ggctcgcacg aaaattcagt ctcagccacg
61 ggtggatccg tgattaagta tttcaacatc aattactaca aggattctgc tagctctggc
121 ttgactaaac aagatttttc ccaagaccca tcgaaattca cacaacctct agcagaagca
181 cttacaaatc cagctttaat gtcaccaact gttgaagcat gtgggatgtc cgataggctt
241 aaacaaatta ctatcgggaa ttccactata acaacacaag atacactaaa ctctatactg
301 gcatatgggg agtggcccaa atacttgagt gacctggacg cttcctcagt ggataagcct
361 acccacccag agacatcatc tgatagattt
//
- Amino acid patent data in Amino acid FASTA format
https://getentry.ddbj.nig.ac.jp/getentry?database=patent_aa&accession_number=BD500001&format=fasta
https://getentry.ddbj.nig.ac.jp/getentry/patent_aa/BD500001?format=fasta
>BD500001|JP 2000316586-A/3: Recombinant microorganism expressing small rubber particle-bound protein (SRPP).
MAEEVEEERLKYLDFVRAAGVYAVDSFSTLYLYAKDISGPLKPGVDTIENVVKTVVTPVY
YIPLEAVKFVDKTVDVSVTSLDGVVPPVIKQVSAQTYSVAQDAPRIVLDVASSVFNTGVQ
EGAKALYANLEPKAEQYAVITWRALNKLPLVPQVANVVVPTAVYFSEKYNDVVRGTTEQG
YRVSSYLPLLPTEKITKVFGDEAS
- AB601234 in nucleotide fasta format
https://getentry.ddbj.nig.ac.jp/getentry?accession_number=AB601234&format=fasta
https://getentry.ddbj.nig.ac.jp/getentry/na/AB601234?format=fasta
>AB601234|AB601234.1 Ainsliaea faurieana chs gene for chalcone synthase, partial cds, haplotype: 2.
ggaccttgctaaaaacaataagggctcacatgtccttgttgtctgctctgagatcattgc
ttccatttttcgtagaccagataagaaccacattgtcagccaagctctctttggggatgg
agcttctgcgctcattgtgggttcagacccagacttctccaaggaacatccattattcaa
gattgtgtctacaactcagacaatcttacagaacactgaaagggcgatgaacttacaatt
gagggaagaagggttgaccattcacctgcacagggatgtaccccagatgacatcaaagaa
tatagaggaggcattagtgcacatatttttgccactgggcataagagactggaactcg
- AB601234 in xml format
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=AB601234&format=xml
https://getentry.ddbj.nig.ac.jp/getentry/na/AB601234/?format=xml
<?xml version="1.0"?>
<!DOCTYPE INSDSet SYSTEM "INSD_INSDSeq.dtd">
-<INSDSet>
-<INSDSeq> <INSDSeq_locus>AB601234</INSDSeq_locus>
<INSDSeq_length>358</INSDSeq_length>
<INSDSeq_moltype>DNA</INSDSeq_moltype>
<INSDSeq_topology>linear</INSDSeq_topology>
<INSDSeq_division>PLN</INSDSeq_division>
<INSDSeq_update-date>18-MAY-2011</INSDSeq_update-date>
<INSDSeq_definition>Ainsliaea faurieana chs gene for chalcone ......
<INSDSeq_primary-accession>AB601234</INSDSeq_primary-accession>
<INSDSeq_accession-version>AB601234.1</INSDSeq_accession-version>
<INSDSeq_source>Ainsliaea faurieana</INSDSeq_source>
<INSDSeq_organism>Ainsliaea faurieana</INSDSeq_organism>
<INSDSeq_taxonomy>Eukaryota; Viridiplantae; Streptophyta; .......
-<INSDSeq_references>
-<INSDReference>
<INSDReference_reference>1</INSDReference_reference>
<INSDReference_position>1..358</INSDReference_position>
-<INSDReference_authors> <INSDAuthor>Mitsui,Y.</INSDAuthor>
<INSDAuthor>Setoguchi,H.</INSDAuthor> </INSDReference_authors>
<INSDReference_title>Direct Submission</INSDReference_title>
<INSDReference_journal>Submitted (17-NOV-2010) to the DDBJ/.....
-<INSDReference>
------- 以下略 -----
- HE963104 in CDS nucleotide fasta format
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=
HE963104&format=cds
https://getentry.ddbj.nig.ac.jp/getentry/na/HE963104/?format=cds
>HE963104-1|CCJ27876.1|111|<1..111|Streptococcus thermophilus|predicted.....
gggttgtcctgtgatgagggaatgctggcagtaggaggacttggtgctgtaggtggcccg
tggggagctgtcggtggggtgttagtaggtgcagccttatactgtttctaa
>HE963104-2|CCJ27877.1|201|130..330|Streptococcus thermophilus|hypothetical.....
atgaataataaacaacttgaaagatttaaaaaactggatacaaatgcattgtctaatgta
agtggtcaaggctatggtgctcaatgtgttattggtactgccggaatgacgattgtcggt
gcagctttctttggcatcgcaggtgcaggagctggatttgcaggcggtagcacagcattt
tgttatggtacagctgaataa
>HE963104-3|CCJ27878.1|219|686..>904|Streptococcus thermophilus|.....
atggcaactcaaacaattgaaaactttaacacccttaacctcgaaacacttgctagtgtt
gaaggaggtggatgtggttggagaggcgcaggtggagcgactgttcaaggagctatcggg
ggagcgtttggaggtaatgtagttttaccagttgtaggctcagttcctggttatctagct
ggtggtgttctaggtggtgcaggtggtactgttgcctat
- JQ677812 in CDS Amino acid translated FASTA format
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=JQ677812&format=trans
https://getentry.ddbj.nig.ac.jp/getentry/na/JQ677812/?format=trans
>JQ677812-1|AFN26948.1|74|Triticum aestivum (bread wheat) HKT1;5
HLAGYSLMLVYLSVVSGARAVLTGKRISLHTFSVFTVVSTFANCGFVPNNEAMIAFRSFP
GLLLLVMPHVLLGI
- DAD (AB000714-1) in nucleotide sequence FASTA format
https://getentry.ddbj.nig.ac.jp/getentry?database=dad&accession_number=AB000714-1&format=cds
https://getentry.ddbj.nig.ac.jp/getentry/dad/AB000714-1/?format=cds
>AB000714-1|BAA22986.1|663|199..861|Homo sapiens|RVP1
atgtccatgggcctggagatcacgggcaccgcgctggccgtgctgggctggctgggcacc
atcgtgtgctgcgcgttgcccatgtggcgcgtgtcggccttcatcggcagcaacatcatc
acgtcgcagaacatctgggagggcctgtggatgaactgcgtggtgcagagcaccggccag
atgcagtgcaaggtgtacgactcgctgctggcactgccacaggaccttcaggcggcccgc
gccctcatcgtggtggccatcctgctggccgccttcgggctgctagtggcgctggtgggc
gcccagtgcaccaactgcgtgcaggacgacacggccaaggccaagatcaccatcgtggca
ggcgtgctgttccttctcgccgccctgctcaccctcgtgccggtgtcctggtcggccaac
accattatccgggacttctacaaccccgtggtgcccgaggcgcagaagcgcgagatgggc
gcgggcctgtacgtgggctgggcggccgcggcgctgcagctgctggggggcgcgctgctc
tgctgctcgtgtcccccacgcgagaagaagtacacggccaccaaggtcgtctactccgcg
ccgcgctccaccggcccgggagccagcctgggcacaggctacgaccgcaaggactacgtc
taa
- View the PDBs in the amino acid FASTA
https://getentry.ddbj.nig.ac.jp/getentry?database=pdb&accession_number=0-Z&format=seqres&limit=5
https://getentry.ddbj.nig.ac.jp/getentry/pdb/0-Z/?format=seqres&limit=5
>100d_A mol:na length:10 DNA/RNA (5'-R(*CP*)-D(*CP*GP*GP*CP*GP*CP*CP*G
CCGGCGCCGG
>100d_B mol:na length:10 DNA/RNA (5'-R(*CP*)-D(*CP*GP*GP*CP*GP*CP*CP*G
CCGGCGCCGG
>101d_A mol:na length:12 DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(CBR)P*GP*
CGCGAATTCGCG
>101d_B mol:na length:12 DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(CBR)P*GP*
CGCGAATTCGCG
>101m_A mol:protein length:154 MYOGLOBIN
----(以下略)----
filetype(optional):Specify the filetype for the result
| default | text |
|---|---|
| html | HTML(links to ACCESSION, ORGANISM, etc) |
| text | TEXT |
| gz | gz compressed |
The name of the gzip files corresponding to the specified formats are as follows.
| [DNA]flatfile | flatfile.txt.gz |
| [DNA]xml | insd.xml.gz |
| [DNA]fasta | fasta_na.txt.gz |
| [DNA]trans | cds_aa.txt.gz |
| [DNA]cds | cds_na.txt.gz |
| [Protein]flatfile | flatfile.txt.gz |
| [Protein]fasta | fasta_aa.txt.gz |
| [Protein]cds | cds_aa.txt.gz |
example (upper:Get method / lower: smart URL)
- download gz of AK377185-AK378194(1000 entries)
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=AK377185-AK378194&filetype=gz&limit=1000
https://getentry.ddbj.nig.ac.jp/getentry/na/AK377185-AK378194?filetype=gz&limit=1000
Following is shown on the display
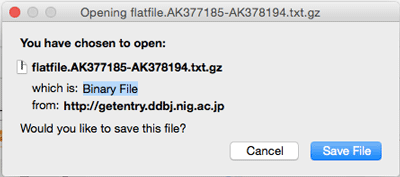
- FW383979 in html
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=FW383979&filetype=html
https://getentry.ddbj.nig.ac.jp/getentry/na/AB601234/?filetype=html
LOCUS FW383979 2675 bp DNA linear PAT 14-OCT-2010
DEFINITION JP 2006521812-A/1: GENETIC POLYMORPHISMS ASSOCIATED WITH RHEUMATOID
ARTHRITIS, METHODS OF DETECTION AND USES THEREOF.
ACCESSION FW383979
VERSION FW383979.1
KEYWORDS JP 2006521812-A/1.
SOURCE Homo sapiens (human)
ORGANISM Homo sapiens
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Mammalia; Eutheria; Euarchontoglires; Primates; Haplorrhini;
Catarrhini; Hominidae; Homo.
REFERENCE 1 (bases 1 to 2675)
AUTHORS Alexander,H.C., Bigobikku,A.B. and Kagiru,M.
TITLE GENETIC POLYMORPHISMS ASSOCIATED WITH RHEUMATOID
ARTHRITIS, METHODS
OF DETECTION AND USES THEREOF
JOURNAL Patent: JP 2006521812-A 1 28-Sep-2006;
APLERA CORPORATION
COMMENT OS Homo sapiens
PN JP 2006521812-A/1
PD 28-Sep-2006
show_suppressed(optional):To display the suppressed data.
| true | display the suppressed data. |
|---|---|
| false | NOT display the suppressed data |
example (upper:Get method / lower: smart URL)
- HE602933 (suppressed)
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=HE602933&show_suppressed=true
https://getentry.ddbj.nig.ac.jp/getentry/na/HE602933?show_suppressed=true
limit(optional): Sets an upper limit of the data acquisition
| default | 10 entries |
|---|---|
| specify the number | specified number of entries |
| 0 | no limit |
example (upper:Get method / lower: smart URL)
- view all results of FY736910 - FY762881(25,972 entries)
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=FY736910 - FY762881&limit=0
https://getentry.ddbj.nig.ac.jp/getentry/na/FY736910 - FY762881?limit=0
trace(optional):When Secondary Accession numberis specified, the result transfers to that of Primary Accession number
| true | display Primary Accession number |
|---|---|
| false | display Secondary Accession number |
example (upper:Get method / lower: smart URL)
- show the primary of AB233943(primary)-AF530906(secondary)
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=AF530906&format=flatfile&trace=true
https://getentry.ddbj.nig.ac.jp/getentry/na/AF530906/?format=flatfile&trace=true
pecifiable parameters in gethistory
accession number {#gh_accession}(mandatory):Specify the accession number for searching.Specification method is the same as getentry
example (upper:Get method / lower: smart URL)
- history search of AB628096
https://getentry.ddbj.nig.ac.jp/gethistory?database=na&accession_number=AB628096
https://getentry.ddbj.nig.ac.jp/gethistory/na/AB628096
AB628096
1 2012-05-25 12:00:00 release 2012-05-25 12:00:00 release live
1 2012-02-24 07:02:55 2012-02-24 07:02:55 live
1 2011-11-25 11:27:22 release 2011-11-25 11:27:22 release live
1 2011-10-22 23:01:47 2011-10-22 23:01:47 live
1 2011-08-26 10:33:50 release 2011-08-26 10:33:50 release live
1 2011-05-27 12:38:45 release 2011-05-27 12:38:45 release live
1 2011-05-11 23:09:49 2011-05-11 23:09:49 live
database(optional):Specify the database for searching.
| default | na |
|---|---|
| DNA | na |
example (upper:Get method / lower: smart URL)
- history search of WGS( BAET01000001)
https://getentry.ddbj.nig.ac.jp/gethistory?database=na&accession_number=BAET01000001
https://getentry.ddbj.nig.ac.jp/gethistory/na/BAET01000001
BAET01000001 BAET01000001
1 2015-09-15 16:20:47 2015-09-15 16:20:47 live
1 2014-06-28 09:14:29 2014-06-28 09:14:29 live
1 2012-06-07 07:05:36 2012-06-07 07:05:36 live
1 2012-06-01 07:05:51 2012-06-01 07:05:51 live
1 2012-03-10 07:10:00 2012-03-10 07:10:00 live
1 2012-03-10 07:03:37 2012-03-10 07:03:37 live
1 2012-02-21 07:03:15 2012-02-21 07:03:15 live
filetype(optional):Specify the filetype for the result.
| default | text |
|---|---|
| html | HTML |
| text | TEXT |
example (upper:Get method / lower: smart URL)
- display the result of AB628096 history search in html format
https://getentry.ddbj.nig.ac.jp/gethistory?database=na&accession_number=AB628096&filetype=html
https://getentry.ddbj.nig.ac.jp/gethistory/na/AB628096/?filetype=html
| accession | version | revision | change | state |
|---|---|---|---|---|
| AB628096 | 1 | 2015-05-29 18:00:00 release | 2015-05-29 18:00:00 release | live |
| 2015-02-27 14:00:00 release | 2015-02-27 14:00:00 release | live | ||
| 2014-11-25 13:00:00 release | 2014-11-25 13:00:00 release | live | ||
| 2014-08-29 21:00:00 release | 2014-08-29 21:00:00 release | live | ||
| 2014-05-30 12:00:00 release | 2014-05-30 12:00:00 release | live | ||
| 2014-02-21 12:00:00 release | 2014-02-21 12:00:00 release | live | ||
| 2013-11-29 12:00:00 release | 2013-11-29 12:00:00 release | live | ||
| 2013-08-30 07:00:00 release | 2013-08-30 07:00:00 release | live | ||
| 2013-05-24 12:00:00 release | 2013-05-24 12:00:00 release | live | ||
| 2013-02-22 12:00:00 release | 2013-02-22 12:00:00 release | live | ||
| 2012-11-22 15:00:00 release | 2012-11-22 15:00:00 release | live | ||
| 2012-08-24 12:00:00 release | 2012-08-24 12:00:00 release | live | ||
| 2012-05-25 12:00:00 release | 2012-05-25 12:00:00 release | live | ||
| 2012-02-24 07:17:46 | 2012-02-24 07:17:46 | live | ||
| 2012-02-24 07:02:55 | 2012-02-24 07:02:55 | live | ||
| 2011-11-25 11:27:22 release | 2011-11-25 11:27:22 release | live | ||
| 2011-10-22 23:01:47 | 2011-10-22 23:01:47 | live | ||
| 2011-08-26 10:33:50 release | 2011-08-26 10:33:50 release | live | ||
| 2011-05-27 12:38:45 release | 2011-05-27 12:38:45 release | live | ||
| 2011-05-11 23:09:49 | 2011-05-11 23:09:49 | live |
How to Create Links to DDBJ Entries
You can create links to individual DDBJ entries.
https://getentry.ddbj.nig.ac.jp/getentry?database=database
name&accession_number=accession number&additional parameters
(optional)
https://getentry.ddbj.nig.ac.jp/getentry/database name/accession number
https://getentry.ddbj.nig.ac.jp/getentry/database name/accession
number/?additional parameters (optional)
For example, a link to AB000001 is
https://getentry.ddbj.nig.ac.jp/getentry?database=na&accession_number=AB000001
https://getentry.ddbj.nig.ac.jp/getentry/na/AB000001
For example,a link to BD500001, an entry of the amino acid sequence
originated in the Patent is not available.
https://getentry.ddbj.nig.ac.jp/getentry?database=aa&accession_number=BD500001
https://getentry.ddbj.nig.ac.jp/getentry/patent_aa/BD500001
As a practical case, please crick the following number to see how the
entry can be viewed.
AB000001
BD500001
Creating links to DRA entries is different from the above method. Please refer to the DRA Web site.