Databases and Data Submission Systems
Databases and Data Submission Systems
The table of databases and data submission systems of the Bioinformation and DDBJ Center.
| Database | Description | Registration site |
|---|---|---|
| Annotated/Assembled Sequences (DDBJ) | For flatfile, a counterpart of GenBank (INSDC). |
• NSSS: Nucleotide Sequence Submission System via web form. • MSS: Data submission system for large scale sequences, not suitable for NSSS. • DFAST: An automatic annotation service for prokaryotic genomes. |
| DDBJ Sequence Read Archive (DRA) | For raw sequencing data and alignment information from high-throughput sequencing platforms including NGS (INSDC). | Submission portal D-way |
| BioProject | Research projects (INSDC) | Submission portal D-way |
| BioSample | Biological source materials and samples (INSDC) | Submission portal D-way |
| Genomic Expression Archive (GEA) | Functional genomics data such as gene expression, epigenetics and SNP genotyping array. | Submission portal D-way |
| MetaboBank | A public repository for metabolomics data. | MetaboBank submission form |
| Japanese Genotype-phenotype Archive (JGA) | Individual-level human genetic and de-identified phenotypic data which require controlled-access. | JGA Submission |
Depending on your research purposes and data categories, you need to submit your data to some of the above databases.
Small-scale Nucleotide Sequence Data Submissions
We recommend you to submit your data via web form, NSSS. In the following cases, please use MSS.
- many number of sequences (greater than 100)
- long sequences (greater than 500 kb)
- complex submission containing many features (more than 30).
- WGS, CON, TSA, TLS, HTC, HTG, EST, GSS and STS submissions
Large-scale Nucleotide Sequence Data Submissions
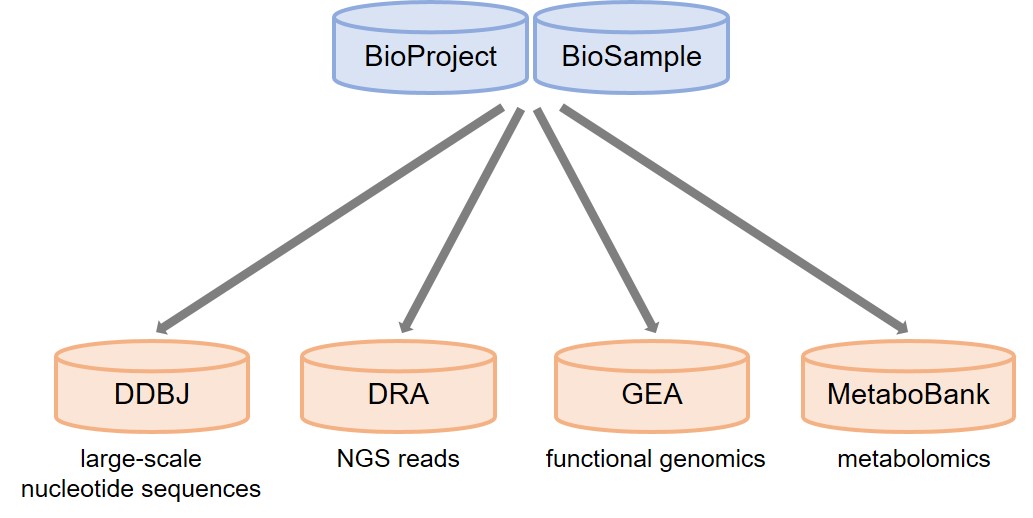
In the following cases, you need to submit your data to DRA and/or MSS after registering BioProject and BioSample.
- Data Submission from Genome Project
- Genome sequences from Bacteria or Archaea, DFAST
- Metagenome assembly
- Single amplified genome
- Data submission from transcriptome project
- Gene expression analysis
- Targeted Locus Study (TLS), large-scale analysis for OTU profiling.
In cases of Transcriptom Shotgun Assembly (TSA), you need to submit your data to both DRA and MSS after registering BioProject and BioSample.
For gene expression analysis by comparative measurements of transcript sequences, you need to submit your data to DRA after registering BioProject and BioSample. We also recommend you to submit processed data to GEA.
Most journals request processed data deposition to GEO/ArrayExpress/GEA.
Biological Data other than Nucleotide Sequences
- We accept microarray data at GEA.
- DDBJ can not accept any amino acid sequences without underlying nucleotide submission. When you want to submit amino acid sequences only, please consider submitting them to UniProt.
FAQ: How to submit amino acid sequences? - In cases of research data from human subjects, we might be able to accept your data at JGA. To submit your data to JGA, a data submission application to DBCLS needs to be approved.
Nucleotide Sequence Data Unacceptable for DDBJ
- Sequence containing a mix of genomic DNA and RNA transcript.
- Sequences without a physical counterpart (consensus sequences).
- Sequences shorter than 100 nucleotides (since June 2021).
- Sequence consisting only of primer (since June 2021).
Submission flow
BioProject/BioSample pre-registration is necessary for large-scale nucleotide sequence submissions to DDBJ as well as DRA/GEA/MetaboBank submissions.