DDBJ Annotated/Assembled Sequences
UME User's manual
UME (Utilities for MSS file Error check) is a stand-alone GUI tool to check syntax and format of Sequence file and Annotation file required for using Mass Submission System of DDBJ and to check amino acid translation of CDS feature based on the description of each file. UME includes both Parser and transChecker.
Basic function
| Name | Functions |
|---|---|
| Parser | Verification of the syntax and format for Sequence file and Annotation file. |
| transChecker | Validation of the amino acid translation for CDS (protein-coding sequence) feature. |
| FFconv | Making simulated format of DDBJ flat file from Sequence file and Annotation file. |
| AGPParser | For the submission of contigs (CON entries), AGP file and Annotation file. |
Install
Download UME package from Validation tool for MSS data file.
Uncompress the downloaded file to extract its contents into the UME folder.
- Windows: Uncompress UME_win.zip
- In case of Windows, you can likely explore the files packed in zip format, directly, like as a folder. However, to run UME, all files in the UME package must be uncompressed.
- unix/macOS: Uncompress UME_unix.zip
- Extract the zipped file using a decompression program or the following command.
$ unzip UME_unix_<Version No.>.zip
- macOS: Regarding the available file names
- DO NOT use multibyte character(s) for any file or folder name of Sequence file, Annotation file, or AGP file when you use UME on some versions of macOS. UME may not work when multibyte character is included in the file or folder name.
Initialization
Starting UME
You can run UME application by double clicking the UME icon (Depending on your PC environment, some extension of UME file would be displayed) in the UME folder.
On macOS, you may not be able to run UME by double clicking the icon.
In that case, please run UME by typing commands on Terminal window like as follows.
$ cd UME # Move to directory that you have expanded UME package.
$ ./UME.sh
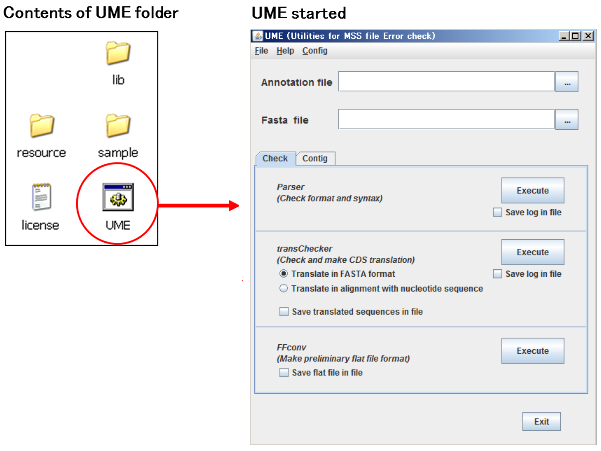
- lib folder
- Directory which includes library files used by UME.
If you move this directory or any file in it, UME would not properly work. - resource folder
- Directory which includes resource files used by UME.
If you move this directory or any file in it, UME would not properly work. - sample folder
- Including sample files for DDBJ submission.
- UME file
- Start UME application by double-clicking this file.
- license file
- A text file for license agreement. Please read at first. The contents of the file are the same as End-user license agreement.
Initial setting of UME
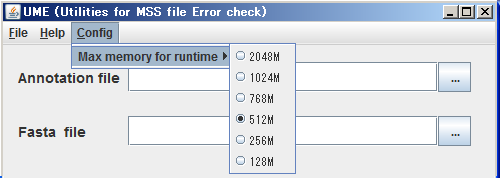
At the first time, you run UME application, change the maximum size of RAM for UME to optimize run-time environments.
The default configuration of it is 512 Mbytes.
(1) Click Config in menu bar.
(2) Select the appropriate size of Max memory for runtime from pull-down menu.
Verification of submission files
Check panel: basic verification of submission files
You can move to this panel by clicking the Check tab enclosed in the red square.
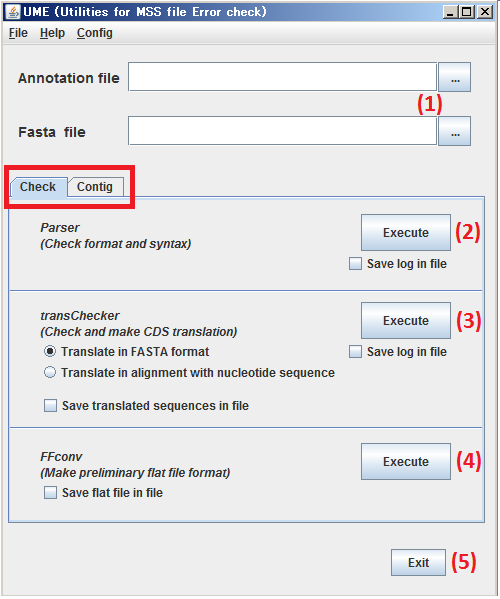
(1) Specify the subjects, both Annotation file and Sequence file.
(2) By clicking Execute button, verify the files specified at step (1) by Parser.
(3) By clicking Execute button, validate amino acid translation of the files specified at step (1) by transChecker.
(4) By clicking Execute button, process the files specified at step (1) by FFconv.
(5) By clicking Exit button, Close UME.
Parser: verification of syntax and format
Execute Parser through the following steps.
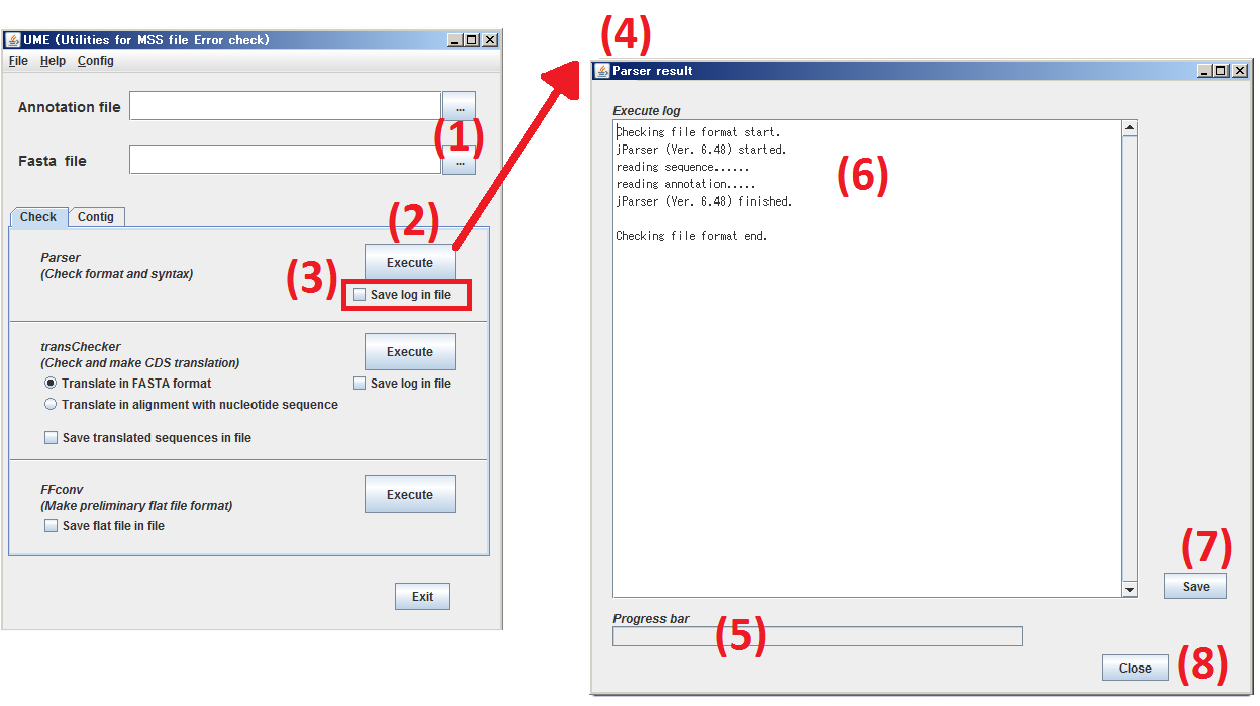
Click Check tab and move to Check panel.
(1) Specify the subjects, both Annotation file and Sequence file.
(2) By clicking Execute button, process the files specified at step (1) by Parser.
(3) When you check the box, Save log in file, the result log of Parser is saved as a text file.
(4) The window of Parser log is opened.
(5) The progress of Parser is indicated.
(6) The result log of Parser is displayed.
(7) By clicking Save button, the contents of (6) are saved as a text file.
(8) By clicking Close button, the window, Parser result, is closed.
When an error occurs, the parser outputs an error message.
Please reffer: Parser Error Eessages, in detail.
transChecker: validation of amino acid translation
transChecker can translate nucleotide sequences of Sequence file into amino acid sequences
corresponding to CDS features specified at Annotation file.
Before using transChecker, both two files should be checked by Parser, and remove all of Parser errors.
Please note that when some Parser error is found, transChecker is terminated without any result. Execute transChecker through the following steps.
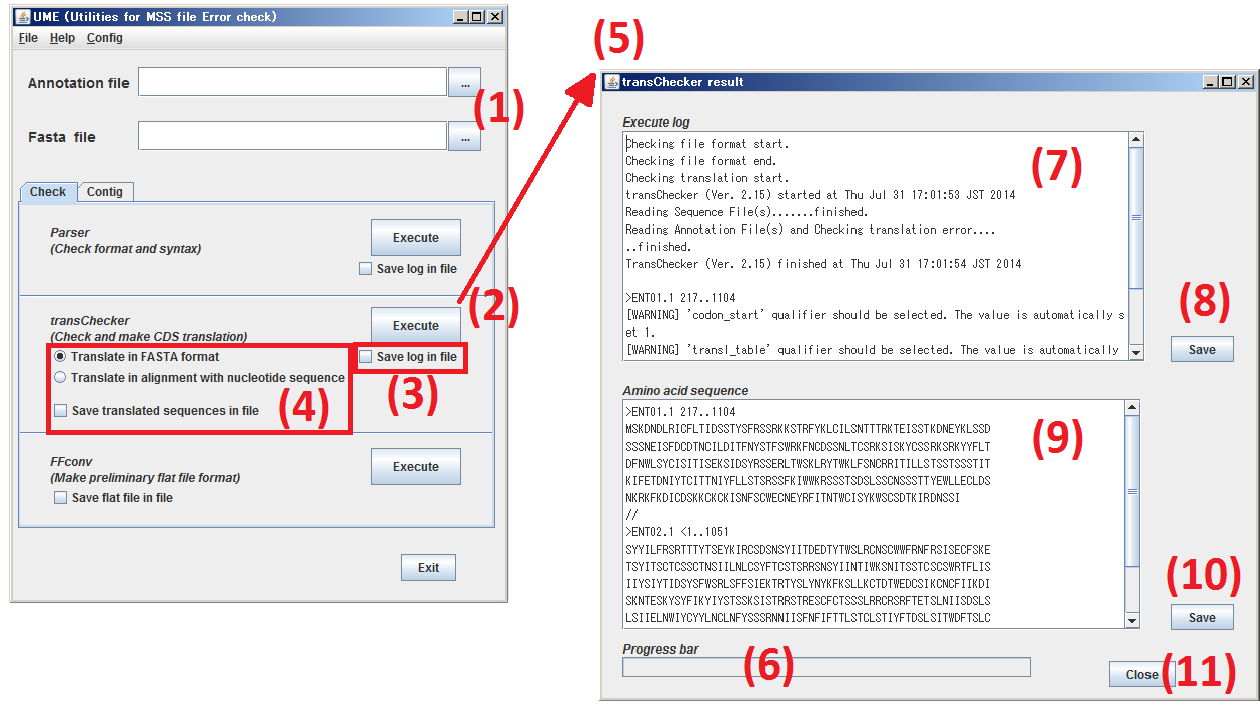
Click Check tab to move to Check panel.
(1) Specify the subjects, both Annotation file and Sequence file.
(2) By clicking Execute button, process the files specified at step (1) by transChecker.
(3) When you check the box, Save log in file, the execution log and error messages of transChecker are saved as a text file.
(4) When you check the box, Save translated sequences in file the amino acid sequences translated by transChecker are saved as a text file in either of two formats.
You can specify the format for translated amino acid sequences from following two types.
| [FASTA-like format](/ddbj/transchecker.html#output-1-1) | in case of selecting Translate in FASTA format |
| [Alignment with nucleotide sequence](/ddbj/transchecker-e.html#output-1-2) | in case of selecting Translate in alignment with nucleotide sequence |
(5) The window of transChecker is opened.
(6) The progress of transChecker is indicated.
(7) The execution log and error messages of transChecker are displayed.
(8) By clicking Save button, the contents of (7) are saved as a text file.
(9) The amino acid sequences translated by transChecker are displayed in the format selected at (4).
(10) By clicking Save button, the contents of (9) are saved as a text file.
(11) By clicking Close button, the window, transChecker result, is closed.
When an error occurs, the transChecker outputs an error message.
Please reffer: transChecker Error Eessages, in detail.
FFconv: confirmation by making simulated DDBJ flat file
FFconv can make simulated format of DDBJ flat file from Annotation file and Sequence file.
Before using FFconv, both two files should be checked by , and remove all of Parser errors. Please note that when some Parser error is found, FFconv is terminated without any result.
Execute FFconv through the following steps.
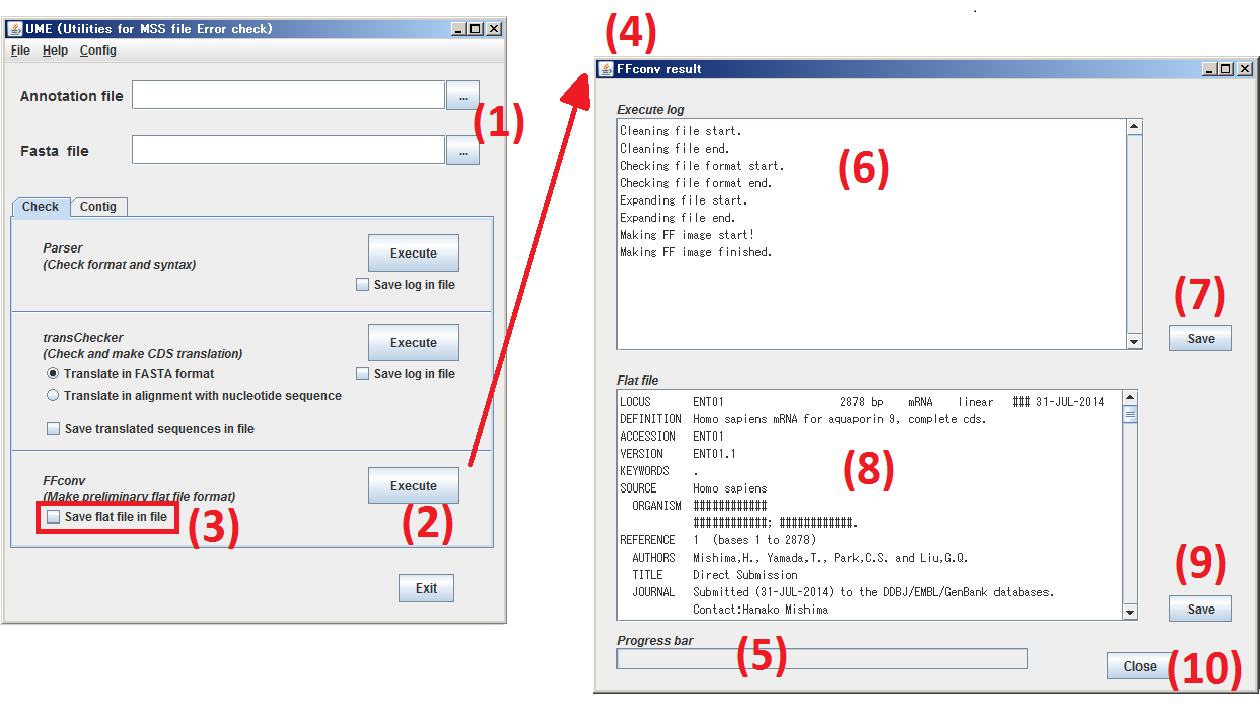
Click Check tab to move to Check panel.
(1) Specify the subjects, both Annotation file and Sequence file.
(2) By clicking Execute button, make simulated format of DDBJ flat file from the files specified at step (1) by FFconv.
(3) When you check the box, Save flat file in file, the result of FFconv, simulated format of DDBJ flat file, is saved as a text file.
(4) The window of FFconv result is opened.
(5) The progress of FFconv is indicated.
(6) The execution log of FFconv is displayed.
(7) By clicking Save button, the contents of (6) are saved as a text file.
(8) The result of FFconv, simulated format of DDBJ flat file, is displayed.
(9) By clicking Save button, the contents of (8) are saved as a text file.
(10) By clicking Close button, the window, FFconv result, is closed.
Verification of contig submission files
[Caution] DDBJ currently terminated accepting new submissions.
Contig panel: verification of AGP file for contig submission
You can move to this panel by clicking the Contig tab enclosed in the red square.
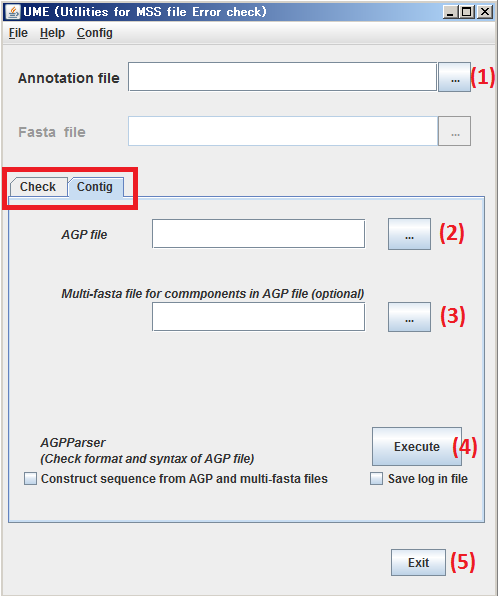
(1) Specify the subject, Annotation file.
(2) Specify the subject, AGP file.
(3) Specify the subject, Multi-fasta file for components in AGP file.
(4) By clicking Execute button, process the files specified at steps (1) to (2) by AGPParser.
(5) By clicking Exit t button, Close UME.
Executing AGPParser
Basically, AGPParser checks the format of AGP file.AGP file.
When you specify Annotation file and/or multi-fasta file including all of sequences cited by AGP file,
AGPParser also checks the consistency between the two or three files.
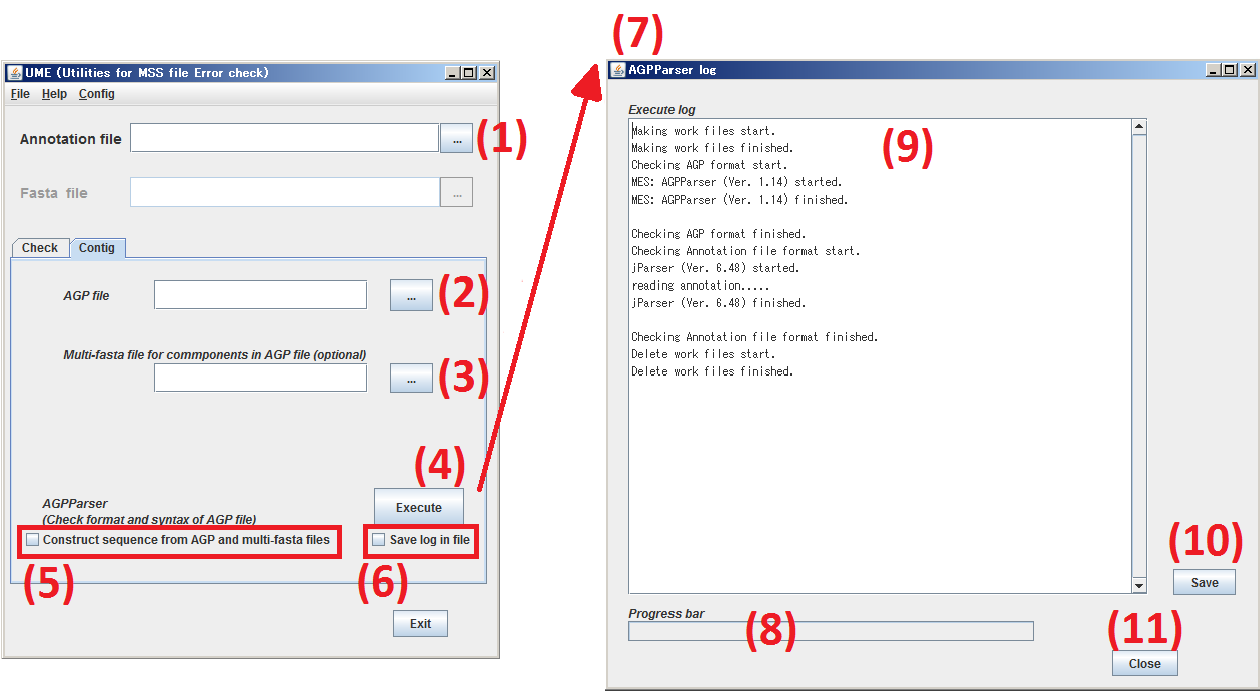
Click Contig tab to move to Contig pannel.
(1) Specify the subject, Annotation file. Not required.
(2) Specify the subject, AGP file. Required.
(3) Specify the subject, the multi-fasta file including all of sequences cited by Sequence file. Not required.
(4) By clicking Execute button,AGPParser checks the specified file at the steps (1) to (3).
(5) When you check the box, [Construct sequence from AGP and multi-fasta files], AGPParser constructs
a text file for nucleotide sequence(s) on accordance with the specification of the two files.
(6) Check the button, Save log in file,then the result log of AGPParser is saved as a text file.
(7) The window of AGPParser log is opened.
(8) The progress of AGPParser is indicated.
(9) The result log of AGPParser is displayed.
(10) By clicking Save button, the contents of (9) are saved as a text file.
(11) By clicking Close button, the window of AGPParser log is closed.
When an error occurs, the AGPParser outputs an error message.
Please reffer: AGPParser Error Messages, in detail.