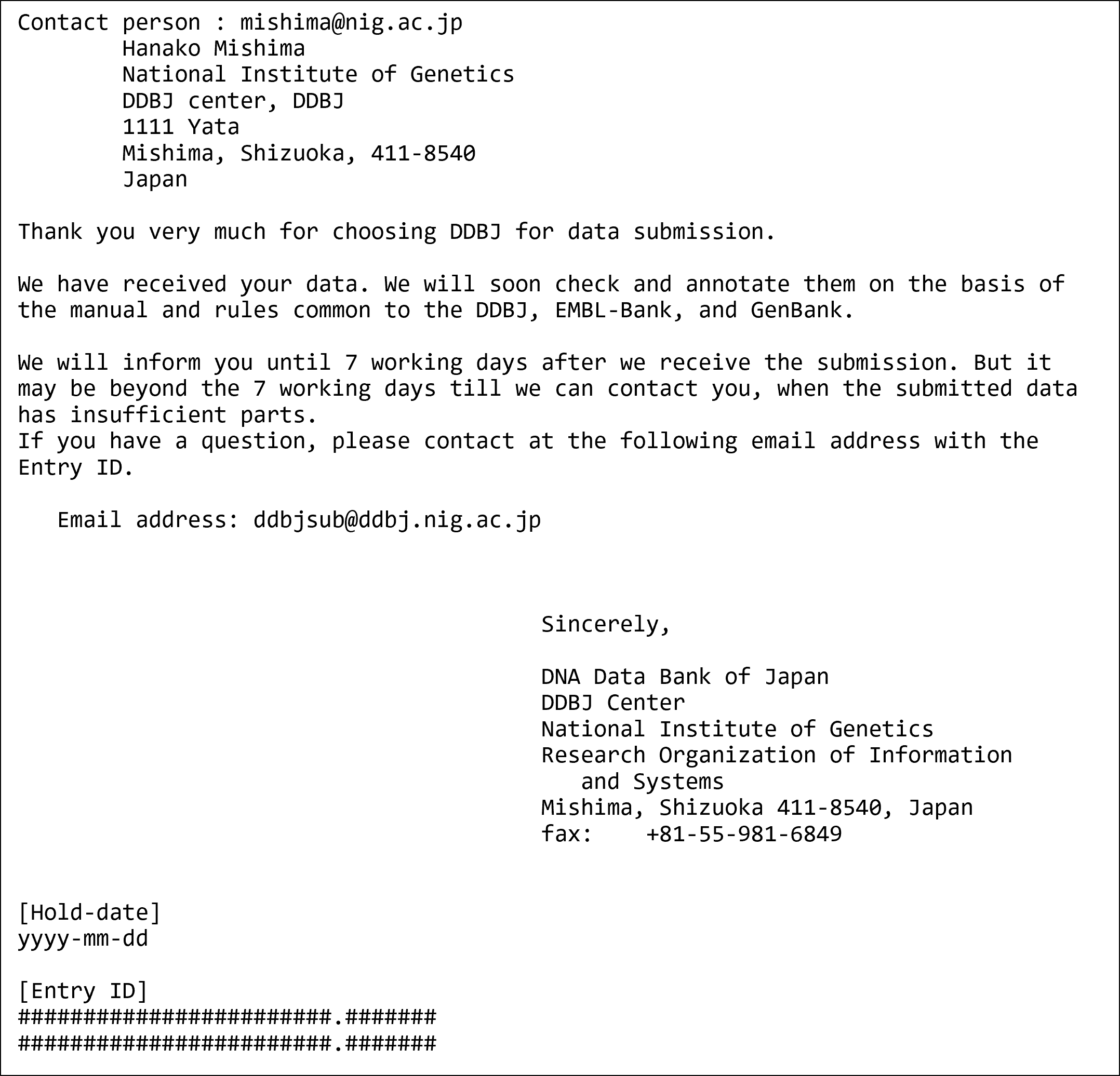
DDBJ Annotated/Assembled Sequences
DDBJ Nucleotide Sequence Submission System HELP
Start
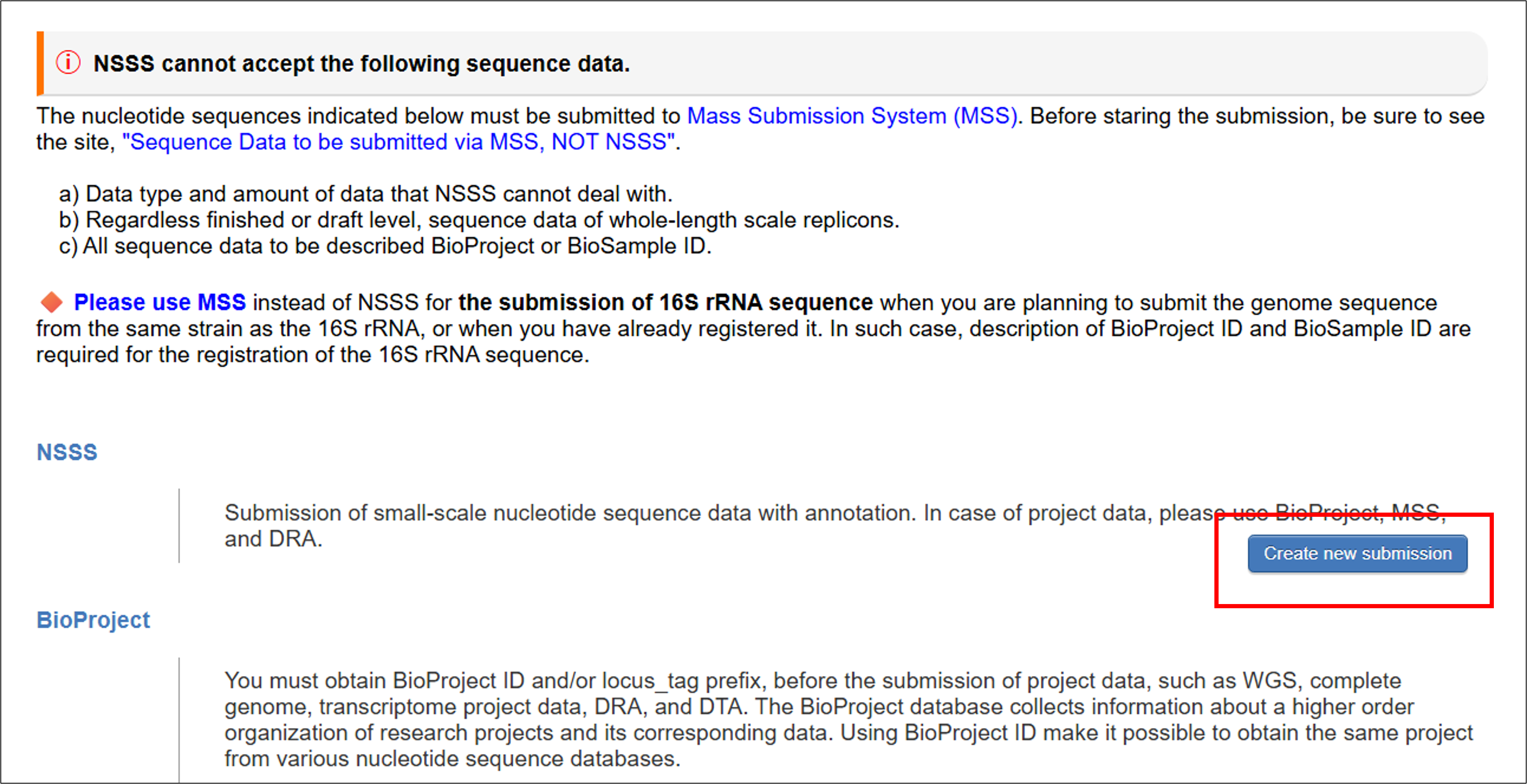
Please open https://ddbj.nig.ac.jp/submission/ and click “Create new submission” to start the submission.
1. Contact person
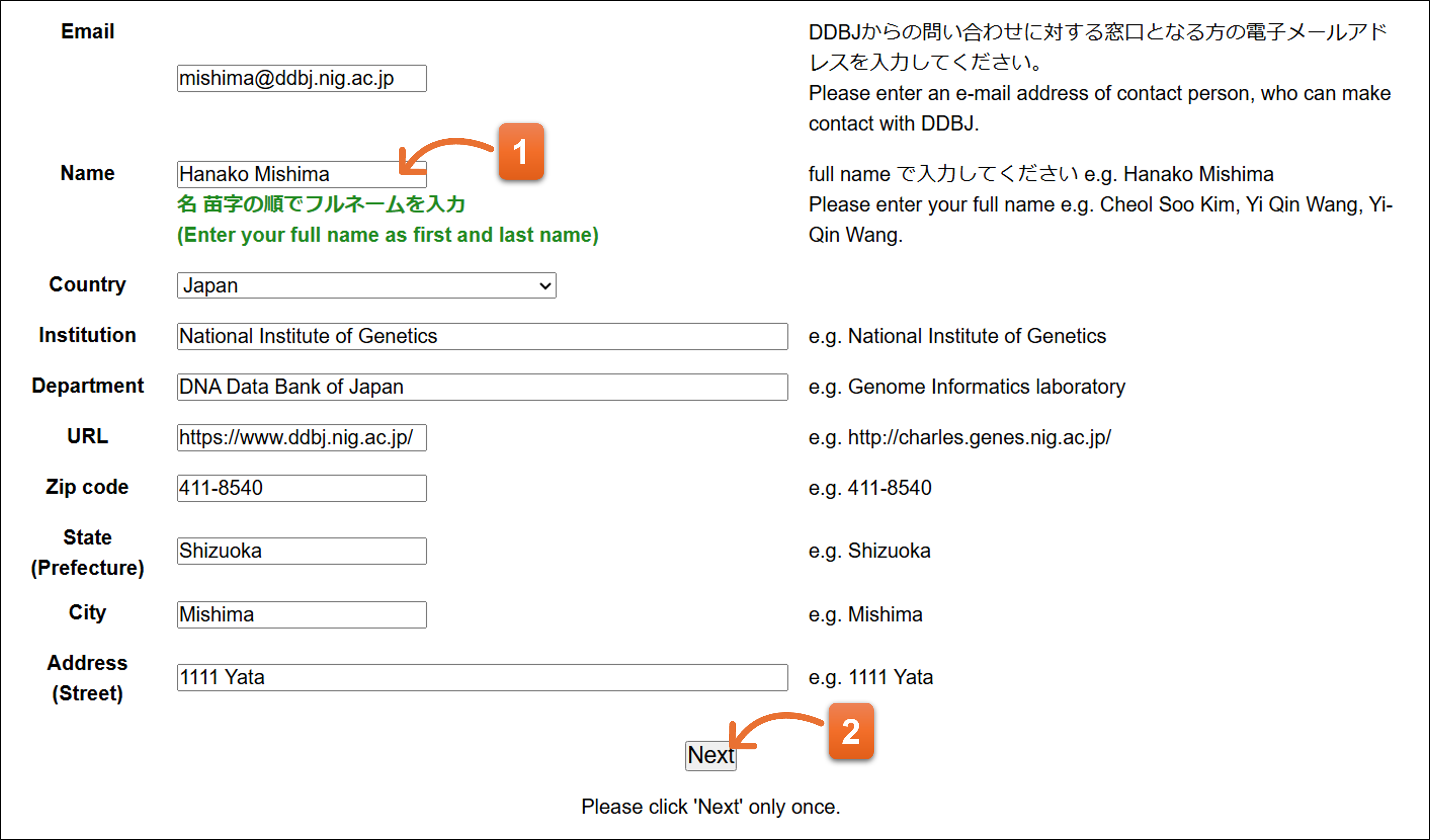
Enter contact person’s information.
- Please enter full name for entering contact person.
- Please click “Next” button after you fill the form. An email will be automatically sent to contact person’s email address.
Please click the hyperlink of the email, then internet browser will open, and you can continue the submission. Or copy the URL to the browser address bar and press enter key.

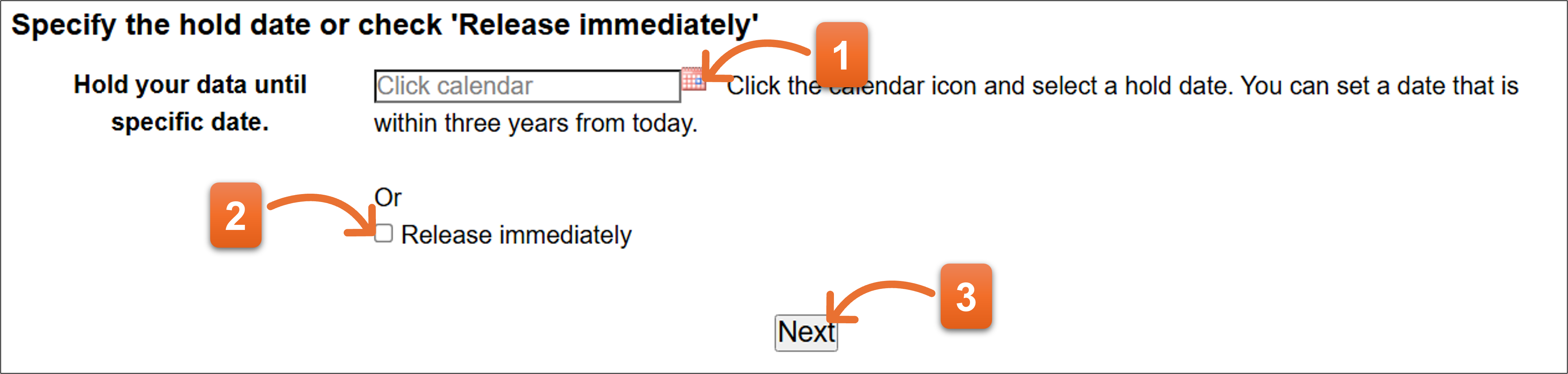
2. Hold date
Enter Hold date if you would like to suspend the release or select “Release immediately” to release right away. You cannot select the days on end or begin of the year because DDBJ suspends the work to release the nucleotide sequences during the days. The selectable hold date is limited within three years from today.
- Click the calendar icon and select date that you intend to withhold from public view.
- Or add the check if you would like to open the nucleotide sequence immediately.
- Click “Next” after you select the hold date.
How to suspend/resume
- On or after the page, you can resume the submission from the bookmarked URL after you click “Next” even if you close the internet browser.
- While you are entering annotation in “7. Annotation” page, entered annotation is saved without clicking the “Next” button. You can resume editing annotation from the bookmarked URL.
3. Submitter
Enter submitter(s) on the page.
- Contact person‘s name is automatically converted to fit the submitter’s format and shown here. You can correct the name if you need to change.
- Click here and you can add more submitter(s). DDBJ recommend that you should include more submitters in the submission. Submitter names on actual nucleotide sequence file are displayed according to the order of this page.
- Click “Next” after you fill the form.
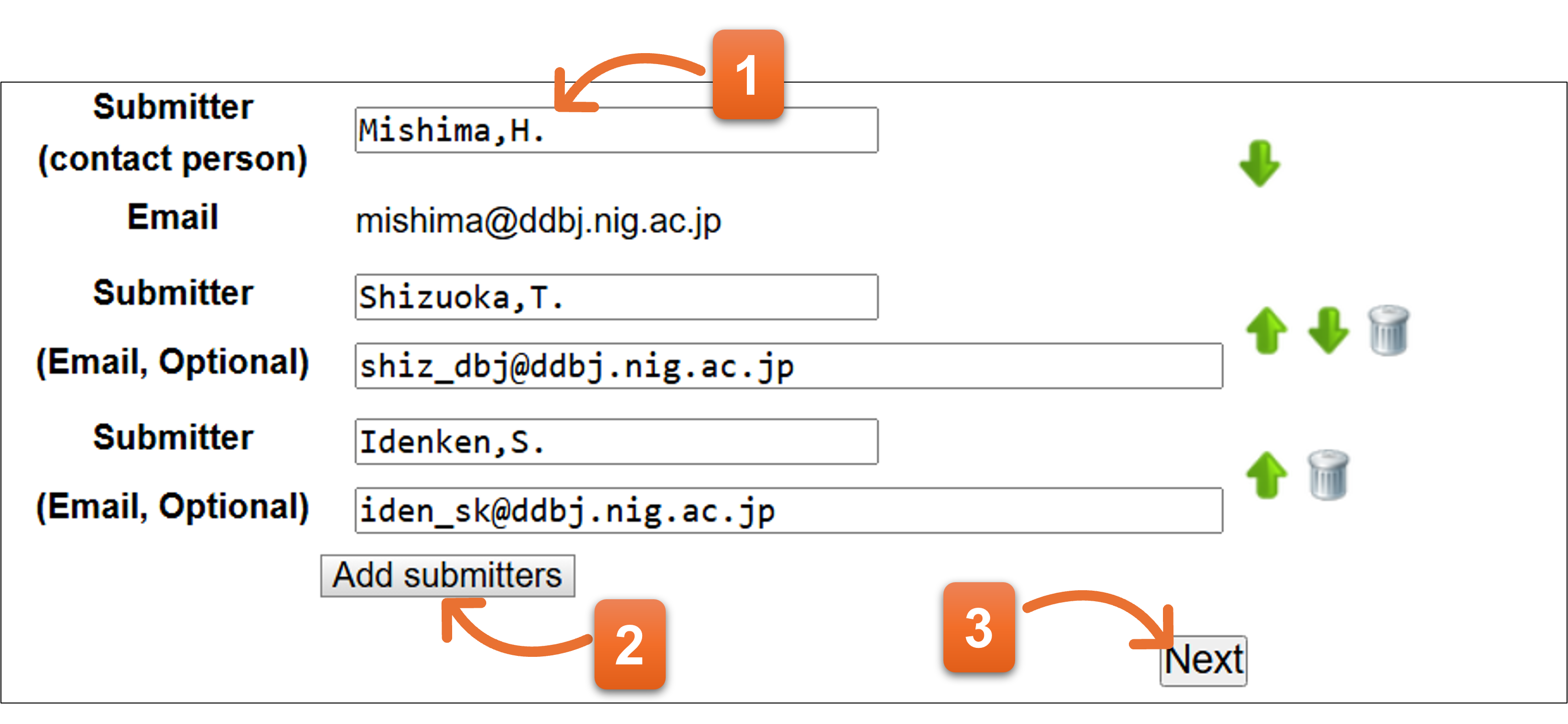
Please enter submitter in abbreviated format as indicated in example below.
Format: Last name[comma]Initial of first name[period]Initial of middle name[period]
e.g.
Miyashita,Y.
Robertson,G.R.
Mishima-Tokai,H.
Kim,C.S.
Wang,Y.Q.
- We would like to ask you to include two or more submitters.
We occasionally meet the situation where we cannot contact to the submitter in case of entries which have only one submitter. In our rule, submitter is responsible for the data and only the submitter can update own entries. Without contacting to the submitter, we cannot fulfill necessary corrections.
Of course, you can register your entries with only one submitter, but we recommend you to add more submitters, such as principal investigator, to your entries.
Related page
DDBJ flat file, REFERENCE 1
4. Reference
Enter reference information on the form. Please enter primary citation on the 1st reference.
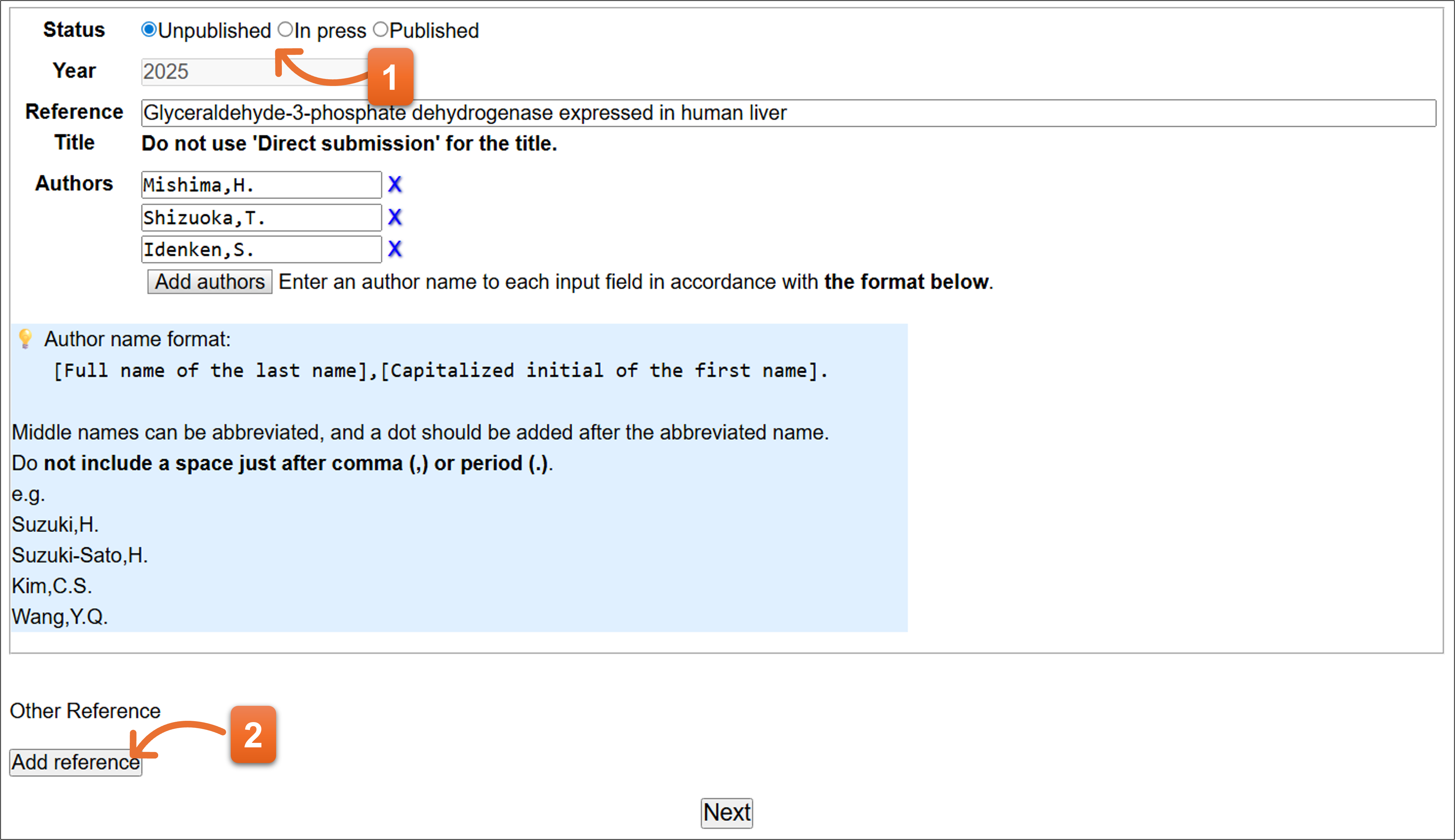
- “Unpublished” is selected as default. Please select “Unpublished” –
when you are planning or preparing for a manuscript or
when a manuscript is being submitted to journal or
when you have not decided to prepare a paper.
Select “In press” when a paper is in press. Select “Published” when aa paper has been published. Text form changes according to a selection at the “Status” - Click if you need to add more reference(s).
Format: Last name[comma]Initial of first name[period]Initial of middle name[period]
e.g.
Miyashita,Y.
Robertson,G.R.
Mishima-Tokai,H.
Kim,C.S.
Wang,Y.Q.
Related page
REFERENCE 2
Sample screenshot on each status
Status: Unpublished
- Current year is automatically filled on the field.
- Enter the paper’s title. Please fill appropriate title even when you are not planning to prepare a paper.
- Please enter reference author (1 person per 1 text box). Click “Add authors” to add more text field. To remove the author, click X icon.
- Click here after you fill the form.
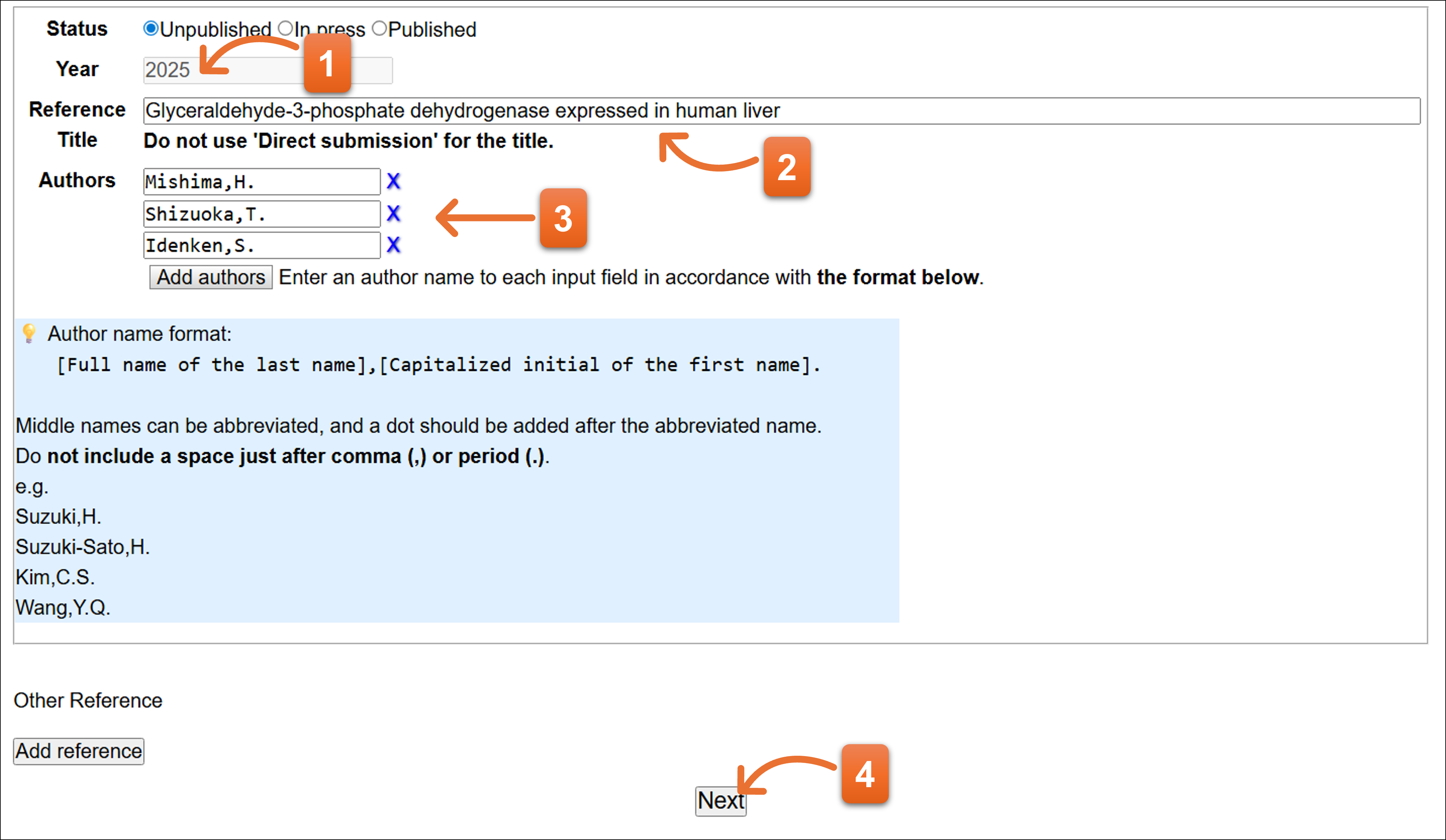
Status: In press
- Enter year.
- Please enter journal abbreviation name. Autocomplete is available here to enter the journal name abbreviation.
- Enter the paper’s title.
- Please enter reference authors (1 person per 1 text box). You can add more text field to click “Add authors”. To remove the author, click “X” icon.
- Click here after you fill the form.
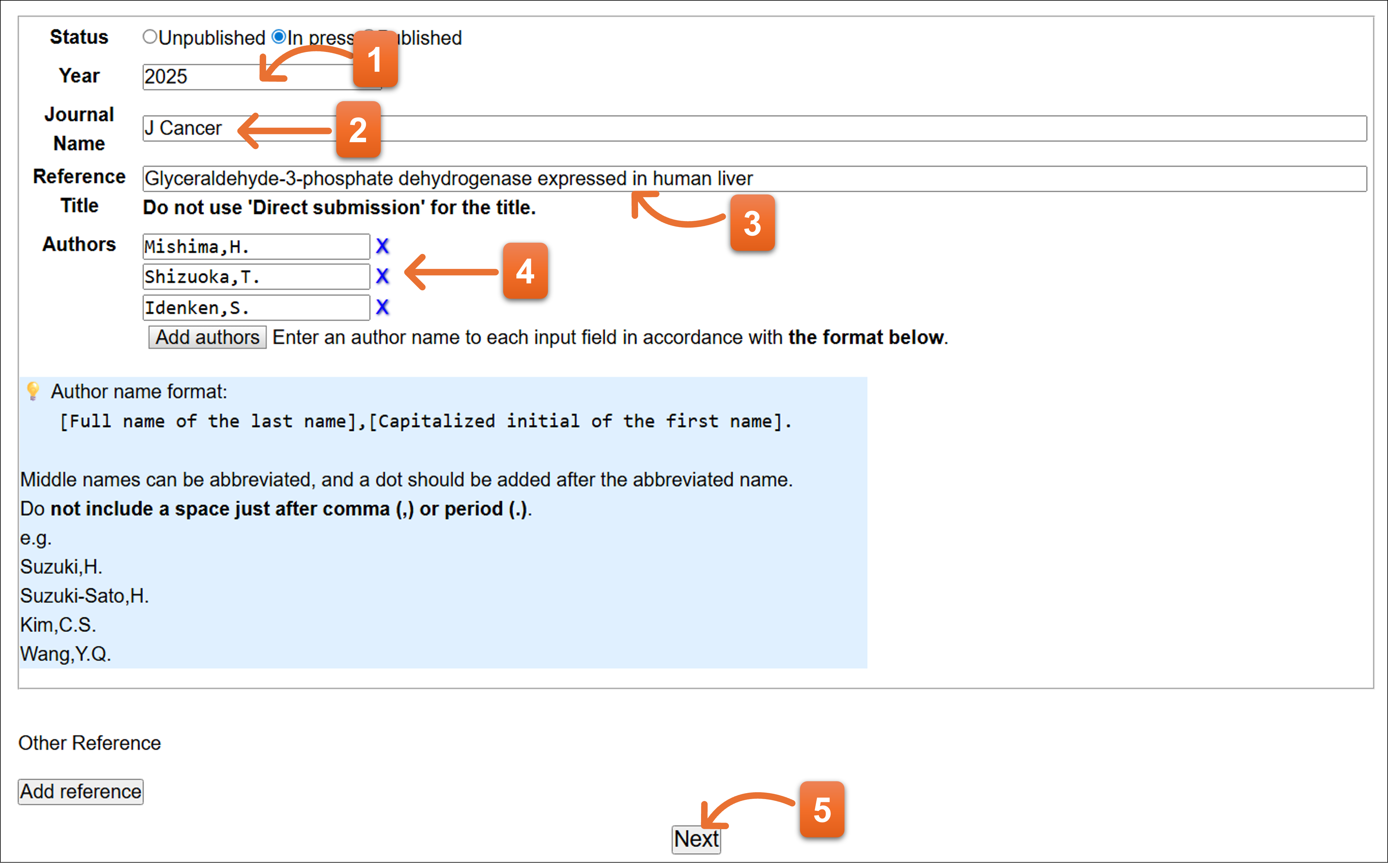
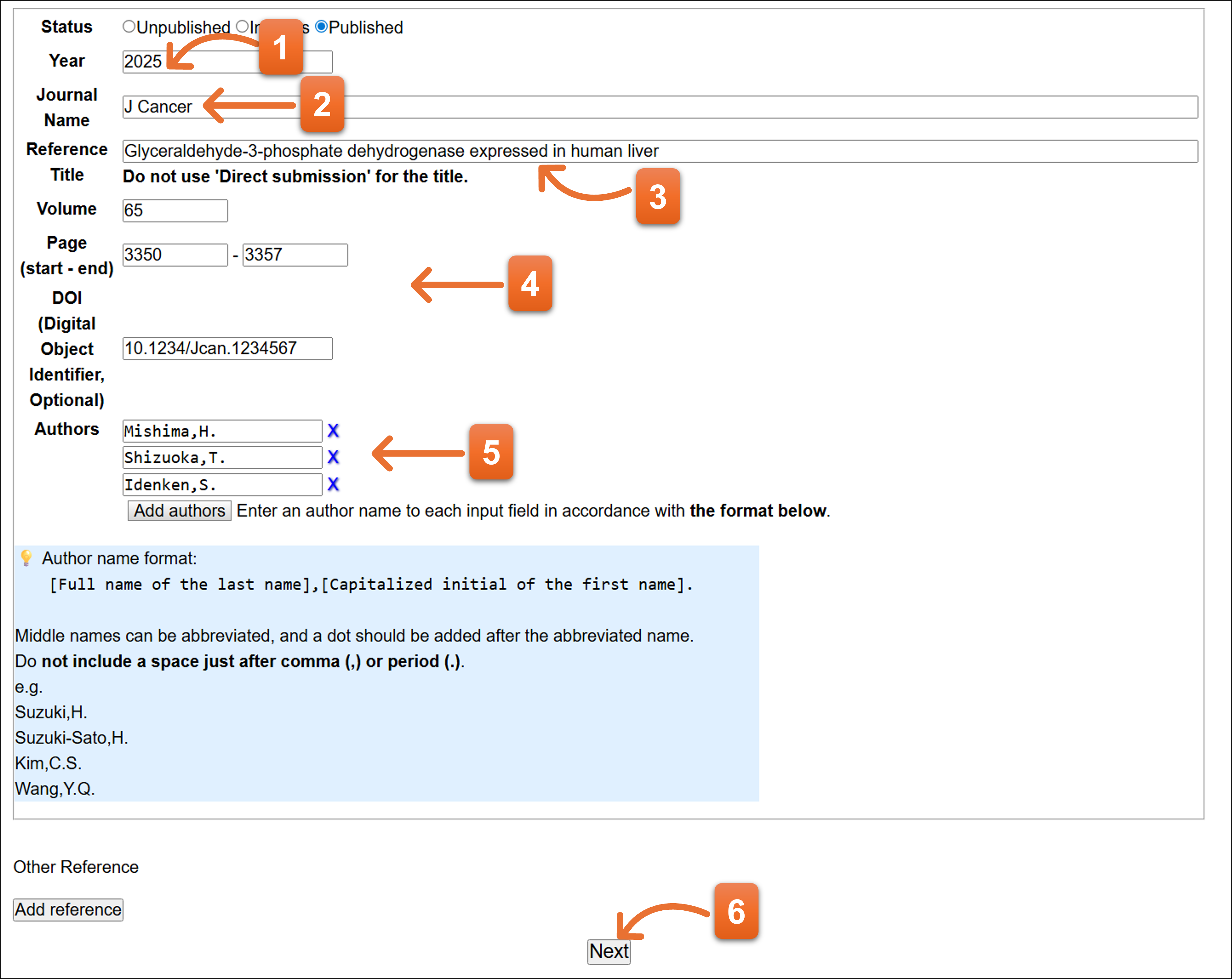
Status: Published
- Enter year.
- Please enter journal abbreviation name. Autocomplete is available here to enter the journal name abbreviation.
- Enter the paper’s title.
- Enter volume, page of the paper. Please fill DOI number if available.
- Please enter reference authors (1 person per 1 text box). You can add more text field to click “Add authors”. To remove the author, click “X” icon.
- Click here after you fill the form.
Journal name
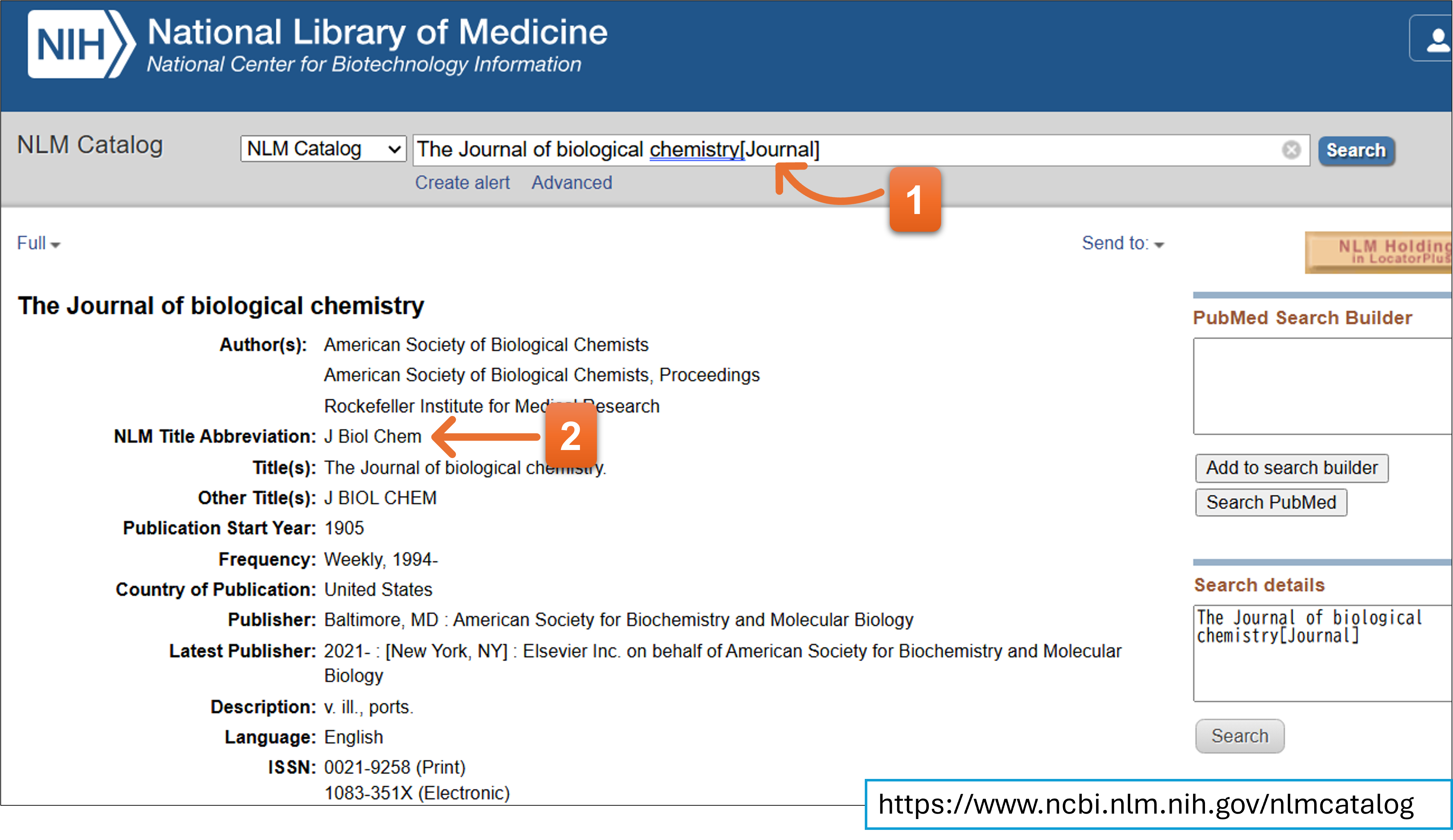
Please enter a journal name in abbreviated format. You will see the candidates of journal name when you enter a full or part of the beginning name of a journal. You can enter the abbreviation name by selecting one from the list.

Regarding abbreviation of the journal name, you can consult it in NLM Catalog
- Search the journal name adding [Journal] just after the query word.
- Please find the “Title Abbreviation” line.
5. Sequence
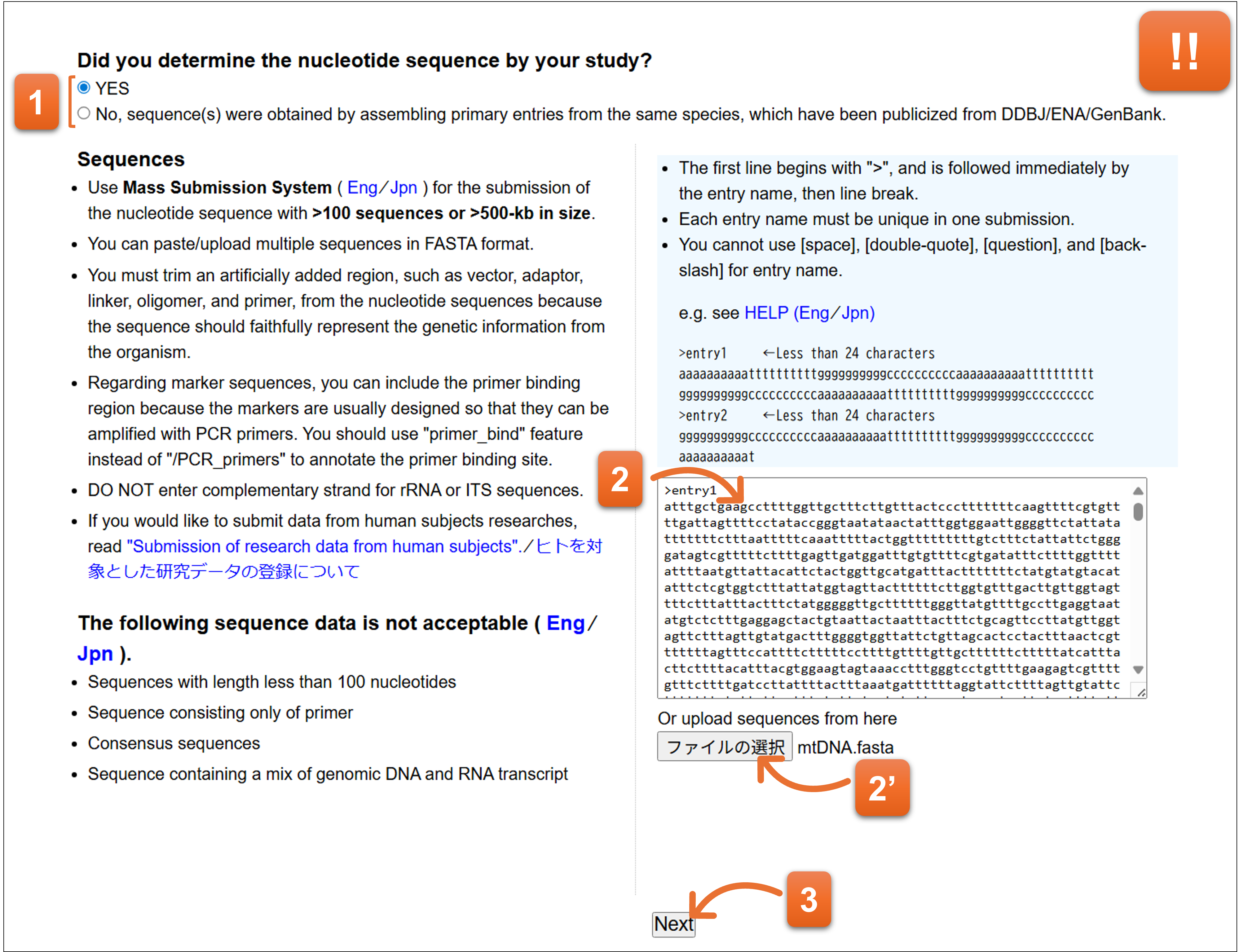
Enter nucleotide sequence here. You cannot submit TPA entry from here. Please go to MSS if you would like to submit TPA.
- “YES” is selected as a default. Please keep “YES” when the nucleotide sequences are not TPA.
- Paste or upload the nucleotide sequences.
- Click here after you fill the form.
📌‼ The information entered at “7. Annotation” form is completely deleted if you change the contents of nucleotide sequence at this form.
Format of the nucleotide sequences
-
You can paste or upload nucleotide sequence consists of multi-FASTA format.
-
Entry name is required to be described in less than 24 letters of characters which do not contain [space], “ [double-quote], ? [question], ¥ [yen sign], \ [back-slash].
-
Entry names must be unique in one submission. If the same entry name are contained in the submission, you must correct the entry name to avoid an error.
-
Double slash (//) is not needed for separate the entries. Of course, you can include double slash (//) as a separation mark of the entries (e.g.1 & e.g.2).
-
This system automatically insert double slash (//) between entries when the nucleotide sequence that contains no double slash (//) is entered.
-
The sequence must consists of a, c, g, t, m, r, w, s, y, k, v, h, d, b, or n.
-
Spaces, numeric characters within the nucleotide sequence are automatically removed.
-
Upper cases of the nucleotide residue id automatically converted into lower cases.
e.g.1
>CLN01
ggacaggctgccgcaggagccaggccgggagcaggtggtggaagacagacctgtaggtgg
aagaggcttcgggggagccggagaactgggccagaccccacaggtgcaggctgccctgtc
tgcgcttcagtcgtgggcgaagcctgaggaaaaagagagagaggctcaaggaagagagga
tgaggcaggagaatcgcttgaaccccggaggcggaggttgcagtgagccgagattacgcc
accgcactccagcctgggcgacagagtgagactccatctcaaaaaaaaaaaaaaaaaa
>CLN02
ctcacacagatgctgcgcacaccagtggttgtaacaatgccgtttgcctccttcaggtct
gaagcctgaggtgcgctcgtggtcagtgaagagggcaaaaagagagagaggctcaaagga
tgcgcttcagtcgtgggcgaagcctgaggaaaaagagagagaggctcaaggaagagagga
tagtcattcatataaatttgaacacacctgctgtgcctagacaagtgtctttctgtaaga
gctgtaactctgagatgtgctaaataaaccctctttctcaaaaaaaaaaaaaaaa
e.g.2
>CLN01
ggacaggctgccgcaggagccaggccgggagcaggtggtggaagacagacctgtaggtgg
aagaggcttcgggggagccggagaactgggccagaccccacaggtgcaggctgccctgtc
tgcgcttcagtcgtgggcgaagcctgaggaaaaagagagagaggctcaaggaagagagga
tgaggcaggagaatcgcttgaaccccggaggcggaggttgcagtgagccgagattacgcc
accgcactccagcctgggcgacagagtgagactccatctcaaaaaaaaaaaaaaaaaa
//
>CLN02
ctcacacagatgctgcgcacaccagtggttgtaacaatgccgtttgcctccttcaggtct
gaagcctgaggtgcgctcgtggtcagtgaagagggcaaaaagagagagaggctcaaagga
tgcgcttcagtcgtgggcgaagcctgaggaaaaagagagagaggctcaaggaagagagga
tagtcattcatataaatttgaacacacctgctgtgcctagacaagtgtctttctgtaaga
gctgtaactctgagatgtgctaaataaaccctctttctcaaaaaaaaaaaaaaaa
//
6. Template
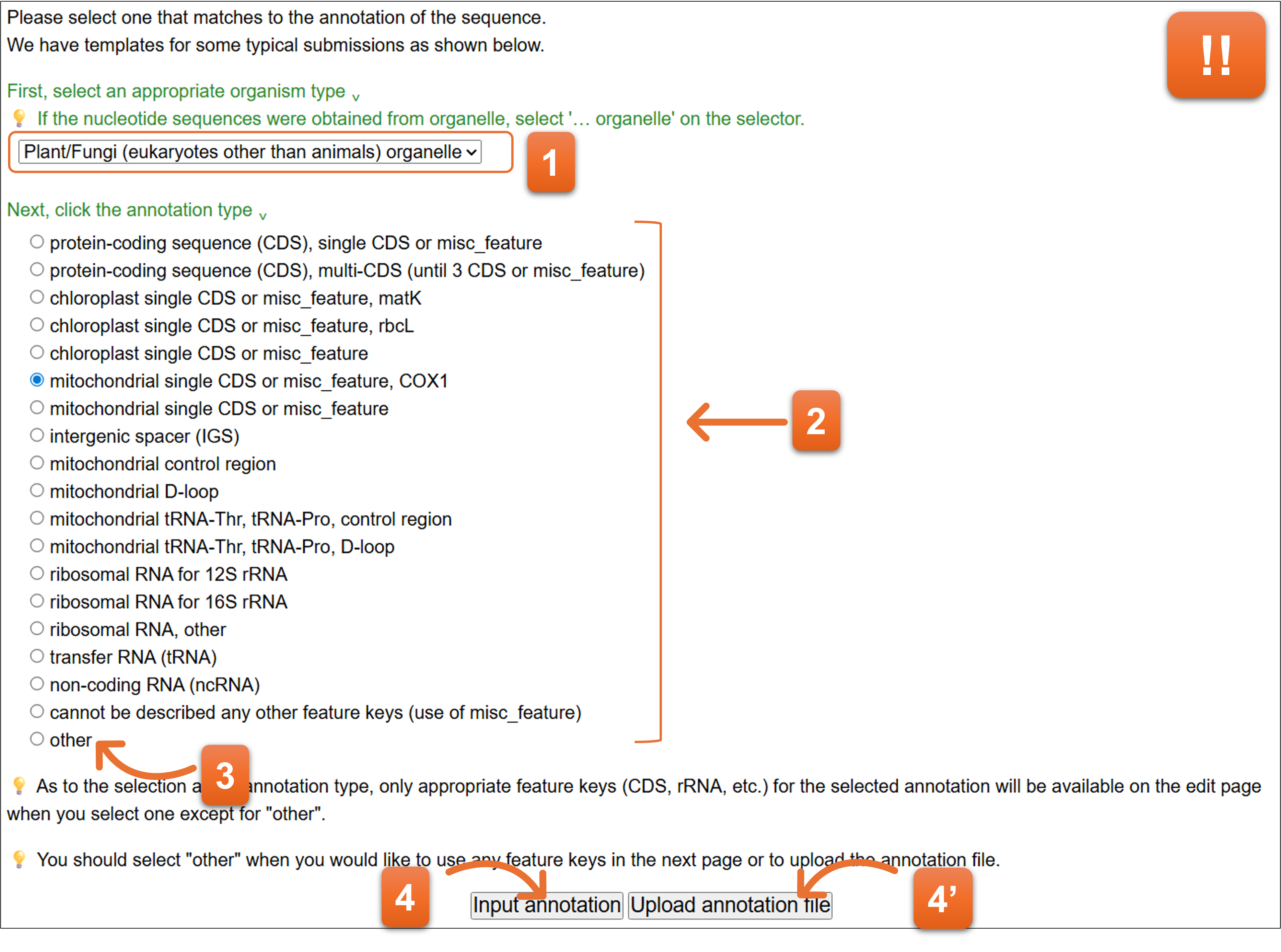
Please select the template that matches to the annotation.
- Please select taxonomic division at first.
- Then select annotation template.
- You should select “other” when you cannot find appropriate annotation in the list or you would like to upload the annotation file. You cannot use spreadsheet type editor for entering annotation when you choose “other”.
- Click “Input annotation” (for entering annotation from Web form) or “Upload annotation file” (for uploading the annotation file) after the selection of template.
📌‼ All information entered at “7. Annotation” are disappeared if you change the template at this form.
7. Annotation
Annotation when a template other than “other” is selected
- Click here to add or remove qualifier keys.
- Click “Edit” icon to enable you edit the annotation per entry.
- You can edit annotation of each cell by double clicking.
- Editing annotation of each column is available from here.
- Please click “Confirm” to check the annotation after you fill entire annotation.
- You cannot click “Next” when error remains after confirming the annotation. You should correct the annotation until there are no errors in the annotation, and then click “Next”.
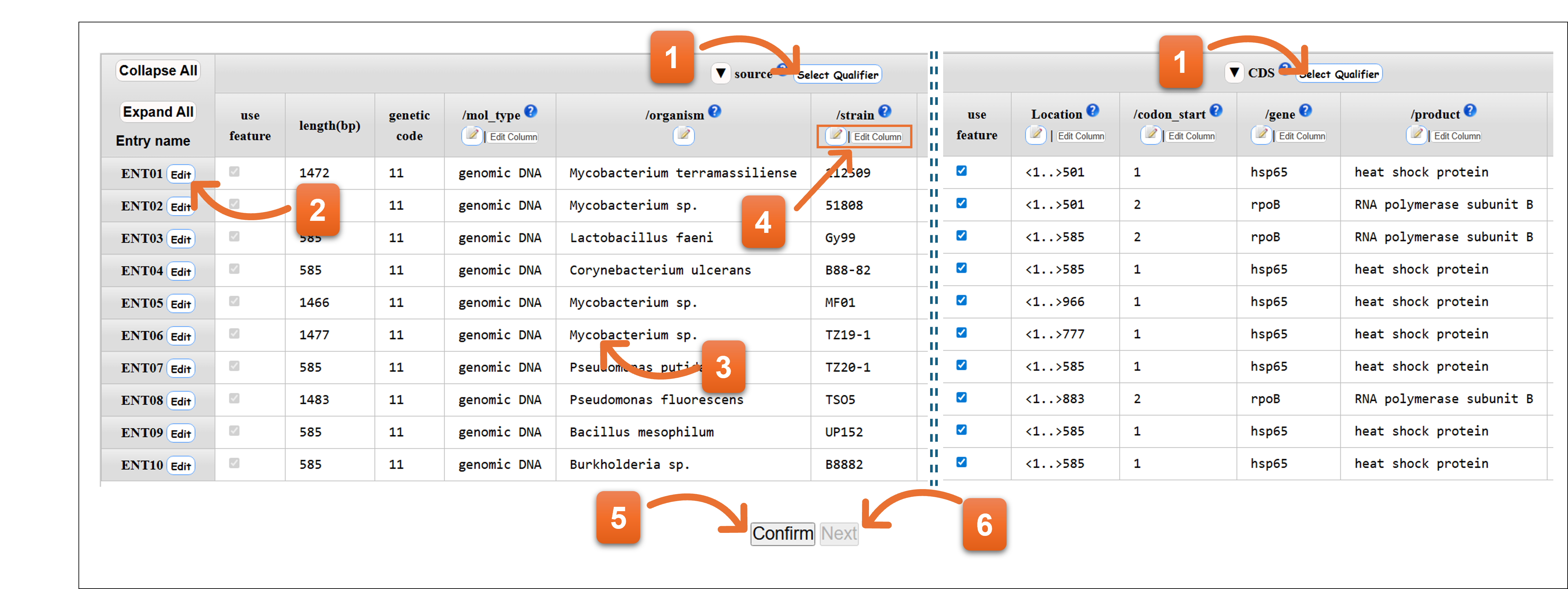
- Related page
- Definition of Feature key / Definition of Qualifier key / Organism qualifier / Protein Coding Sequence; CDS feature
Annotation when template “other” is selected
- Click here to add or remove qualifier keys.
- Click the icon to copy the qualifier keys selected for source feature of this entry to all subsequent entries.
- Please add feature keys from here.
- You can edit annotation by clicking each location and qualifier key.
- Click “Edit” icon to enable you edit the annotation per entry.
- Please click “Confirm” to check the annotation after you fill entire annotation.
- You cannot click “Next” when error remains after confirming the annotation. You should correct the annotation until there are no errors in the annotation, and then click “Next”.
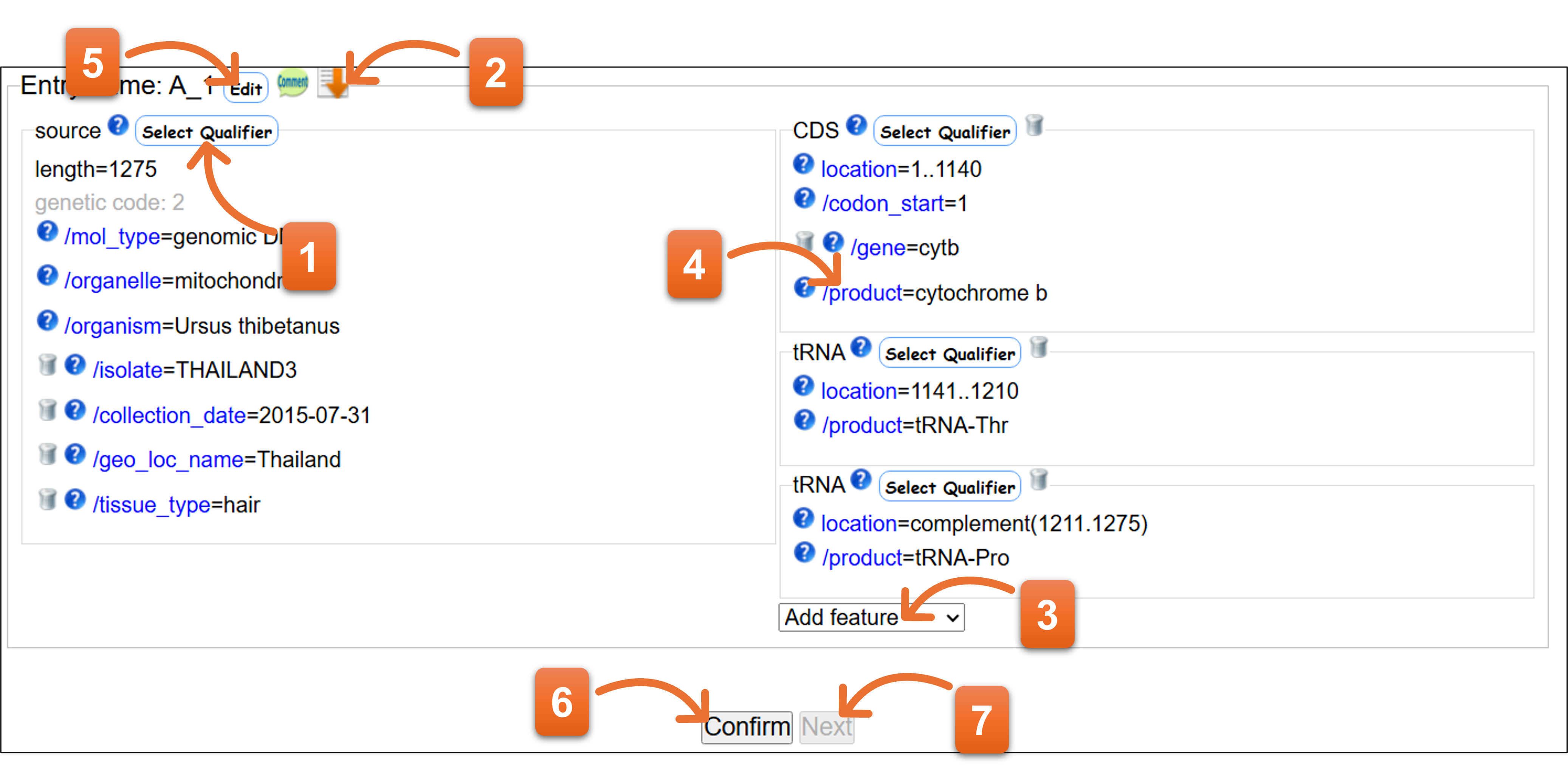
- Related page
- Definition of Feature key / Definition of Qualifier key / Organism qualifier / Protein Coding Sequence; CDS feature
How to edit the annotation
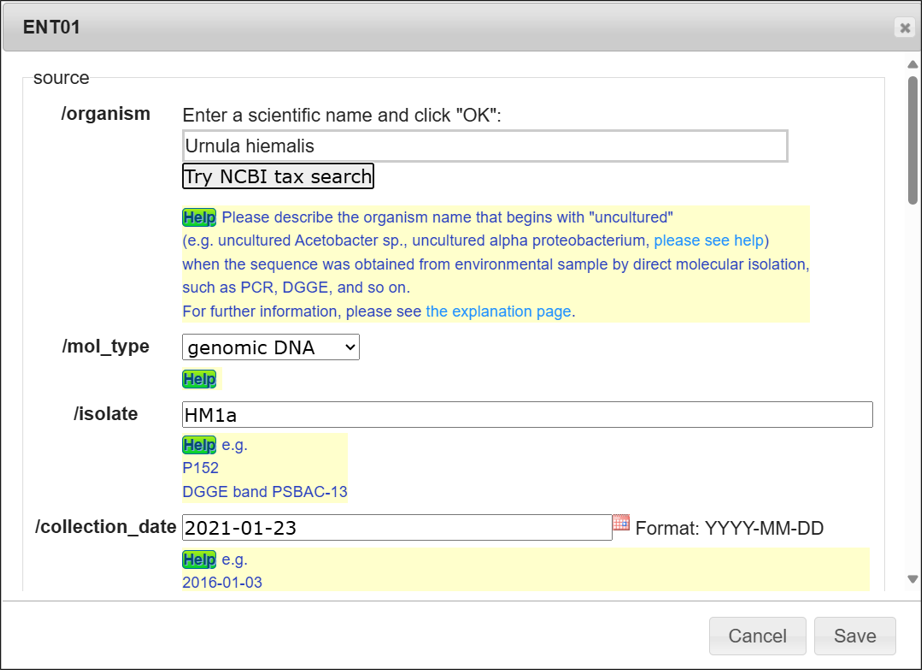
“Edit” button
You can input annotation one-by-one entry. Please click “Save” after you fill the annotation.
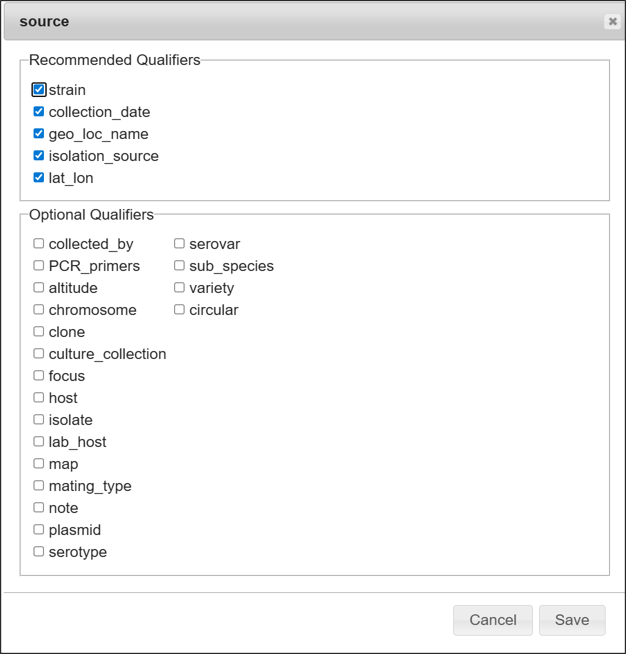
“Select qualifier” button
Adding or removing check mark against the checkbox besides list to add or delete qualifier keys. Available qualifier keys varies according to feature key or the type of annotation template.
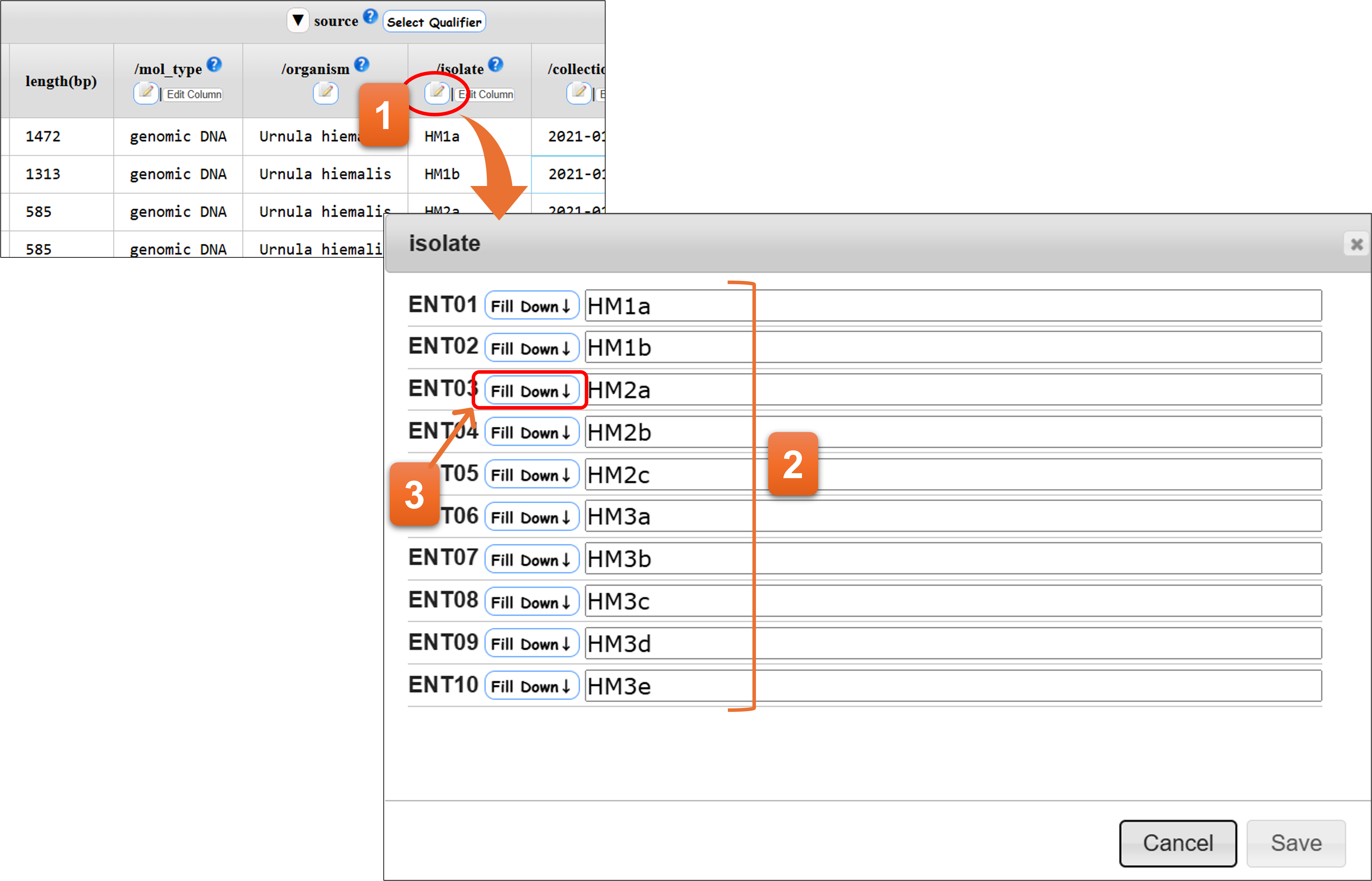
“Pen & Note” button
You can edit qualifier values per column on the annotation table.
- Click here to start editing.
- You can edit the values.
- To copy the same value to subsequent entries, click “Fill Down”.
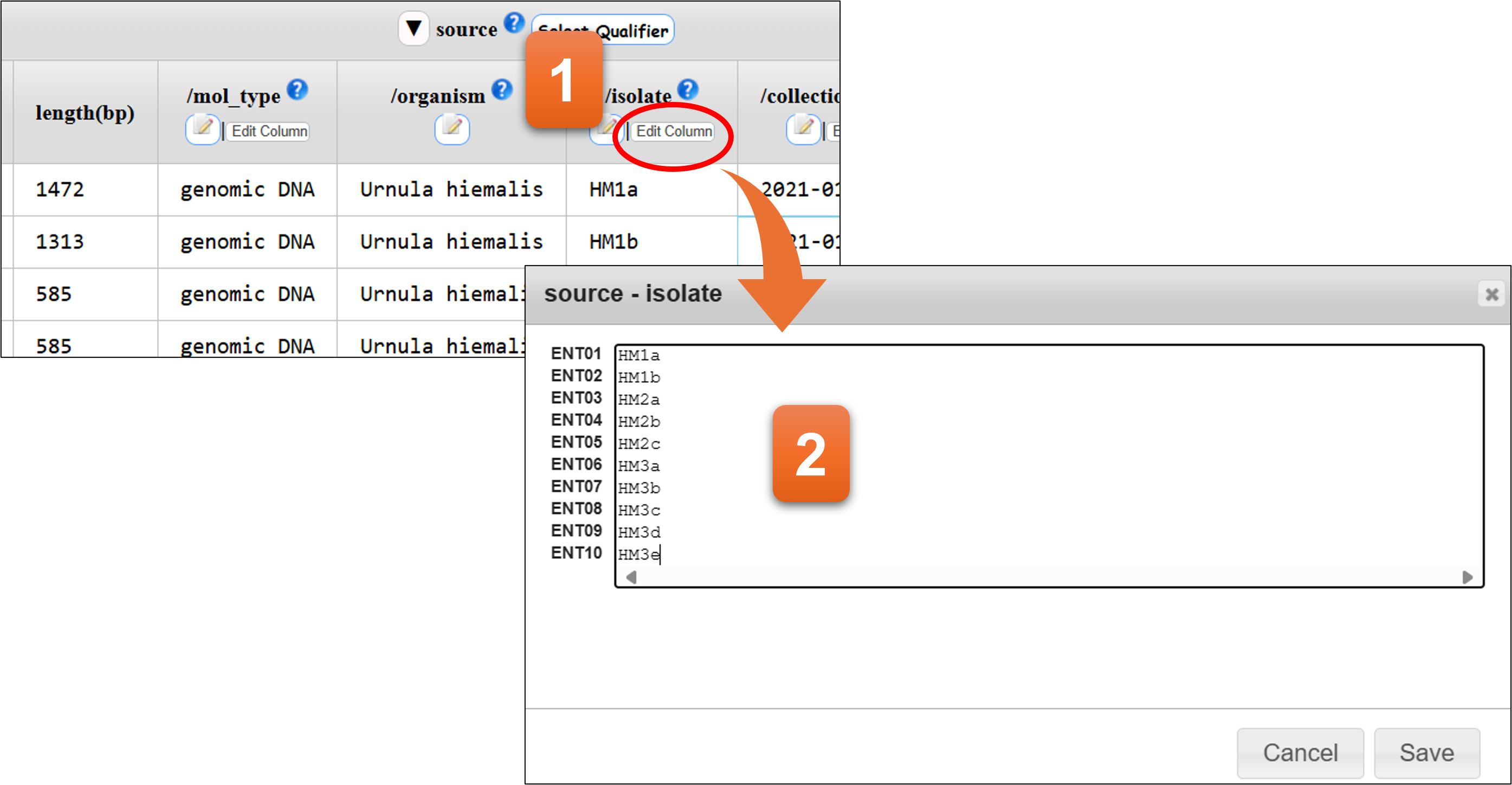
“Edit Column” button
- Click here to start editing.
- You can edit directly on the text area or copy & paste the data from Excel or text editor. Please use line feed to separate each value.
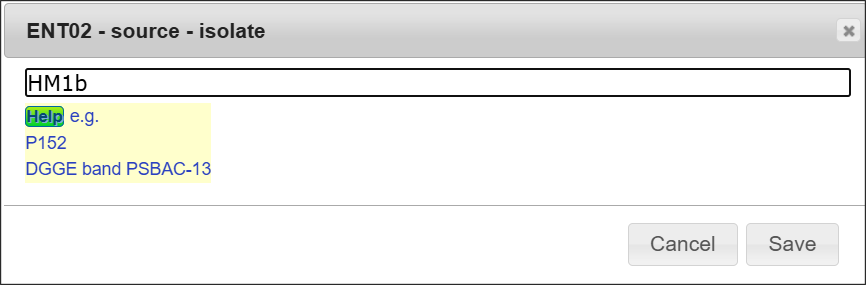
Double-clicking the cell
Double-clicking the cell (or clicking each qualifier/location when template - other is selected) to open the input field. Please enter or edit value against the field.
Organism name
Enter a scientific name here. The inputting system support you to enter a correct scientific name. Candidate names will be displayed on the list while typing organism name, then you can click one from the list and click “OK”.
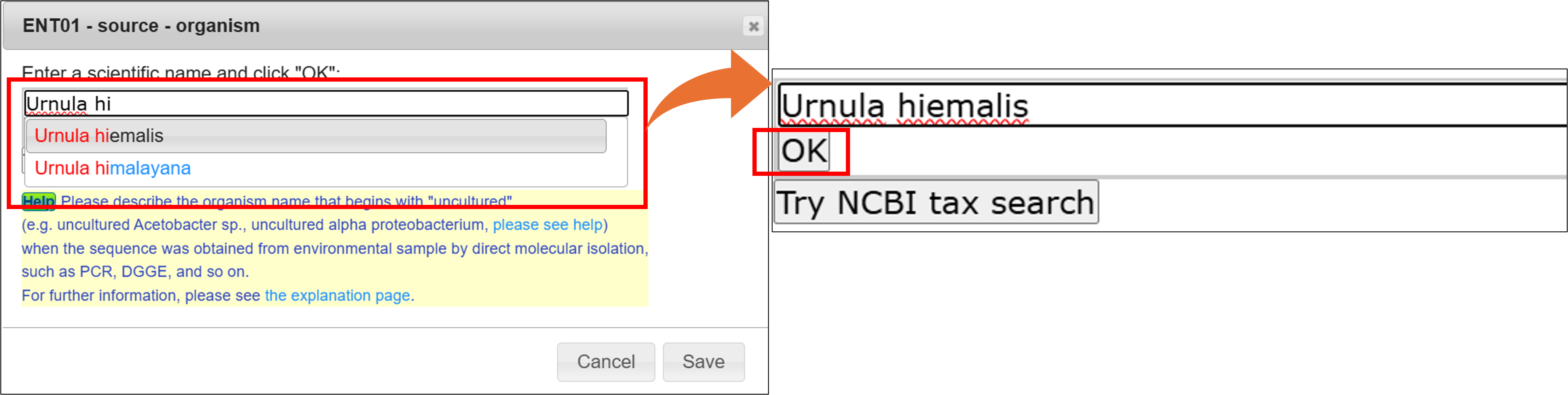
You need to select one from the category list, if the organism name is not registered in NCBI Taxonomy database. Please see Category of organism name for detail.
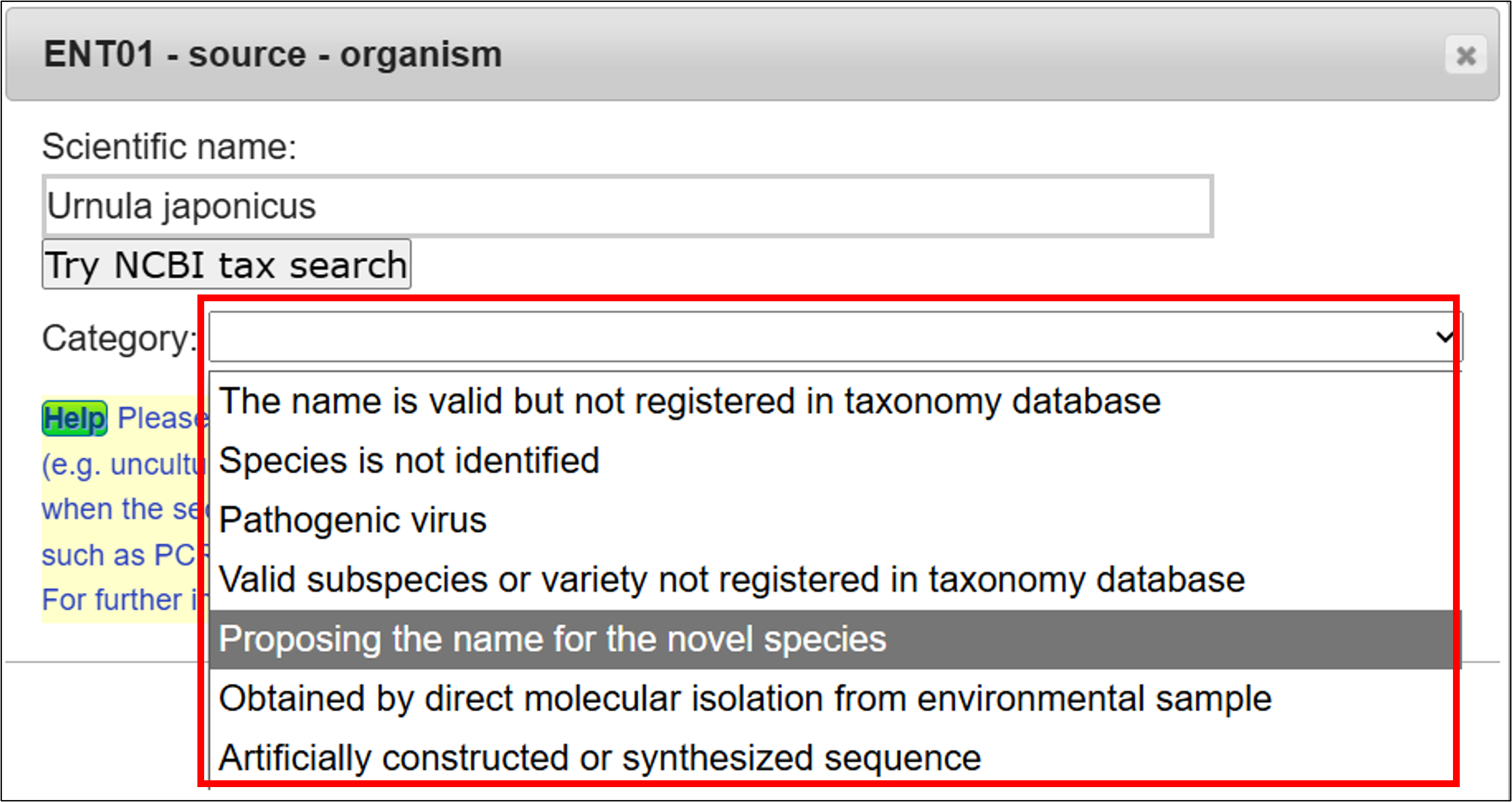
- Related page
- Organism qualifier
Annotation examples
16S rRNA
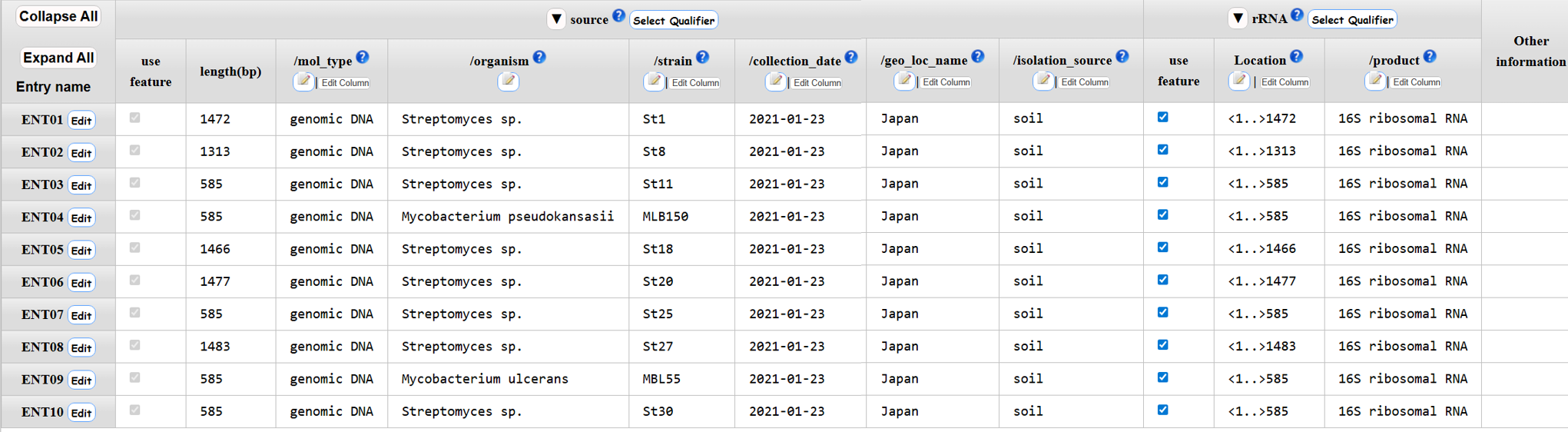
CDS
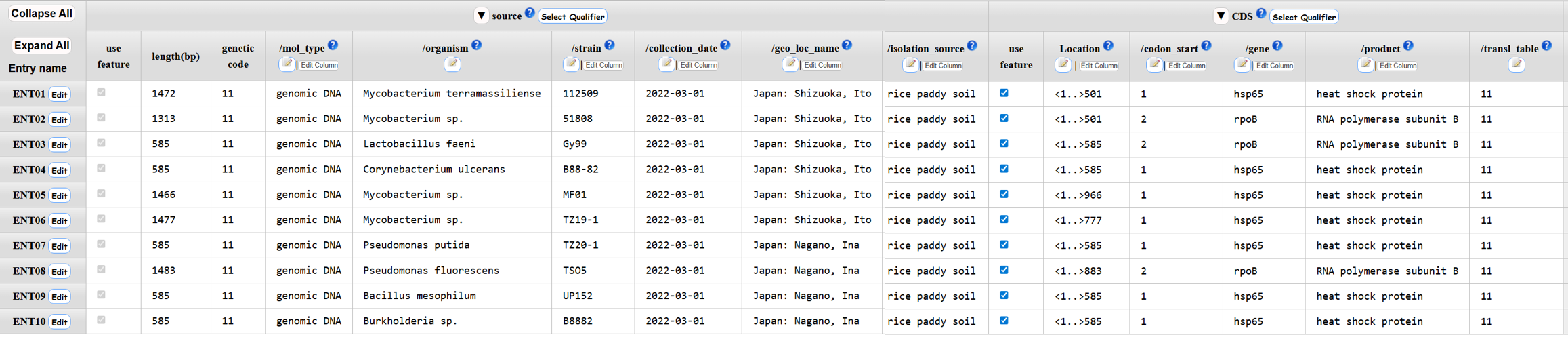
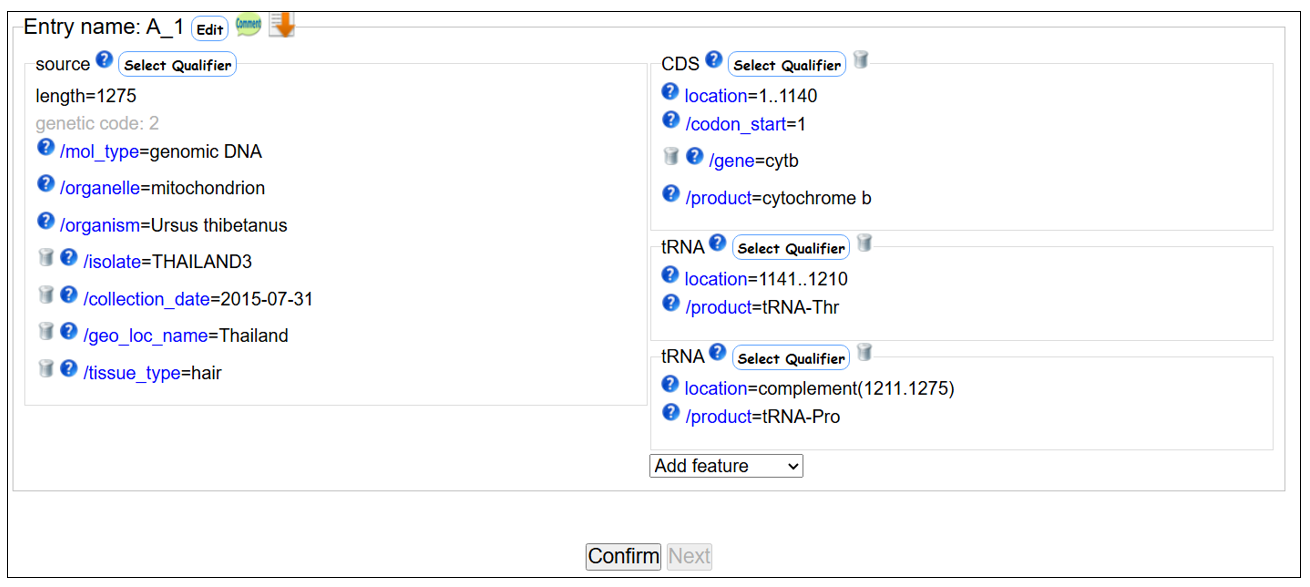
Mitochondrial cytb - tRNA-Pro
Uploading the annotation file
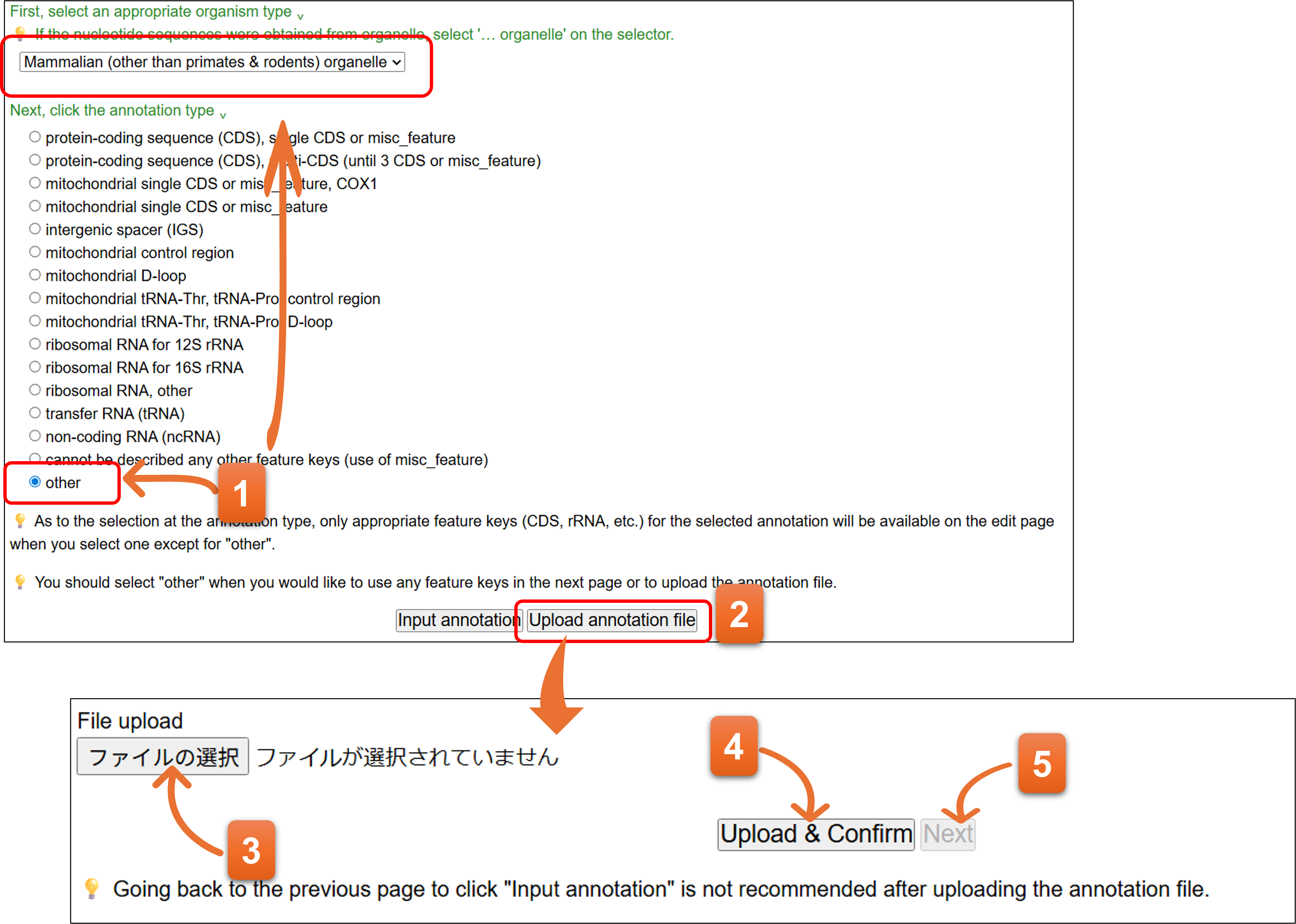
- On the template form, select an appropriate taxonomic division and then select “other” among the annotation type.
- Please click “Upload annotation file”.
- Please set the annotation file by clicking the button.
- Then click “Upload & Confirm” button to check the annotation file.
- You cannot click “Next” when there are any errors in the annotation file. Please correct the annotation file and carry out No.3 and No.4 again. “Next” will be available when there are no errors in the annotation file. Please click “Next” to proceed.
Uploadable file format
-
You can download sample annotation file from here. You should include biological feature in the annotation file.
-
Please refer to “Making MSS Files for preparation of annotation file” for detail.
-
You cannot upload any one of the annotation for WGS, TSA, TLS, EST, HTC, GSS, STS, HTG, or TPA, nor the file containing DBLINK and ST_COMMENT. Please contact “Mass Submission System (MSS)” to submit such submission files.
-
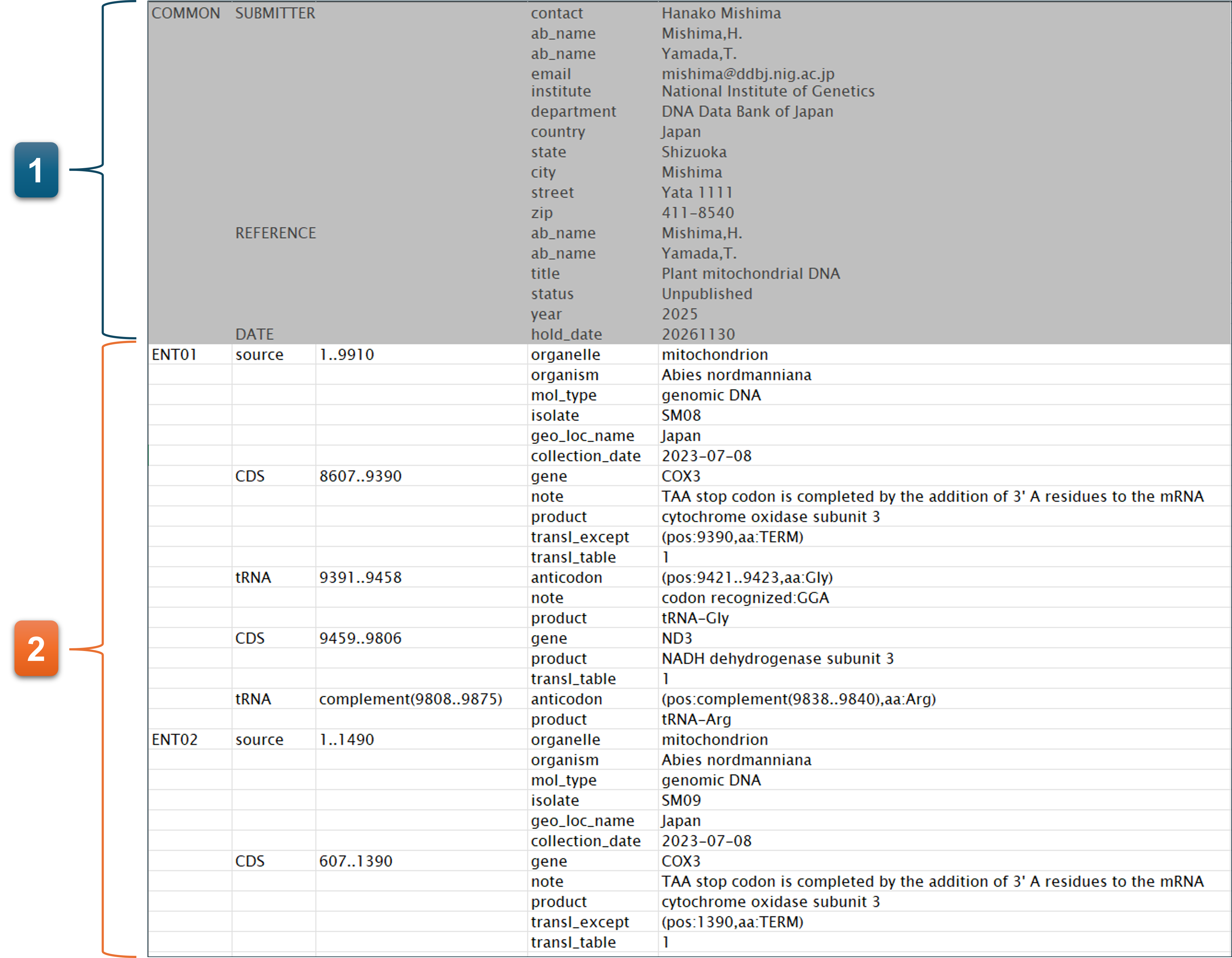
Information that you entered on the pages, “1. Contact person”, “2. Hold date”, “3. Submitter”, and “4. Reference”, are automatically added in front of uploaded annotation file as COMMON section.
-
When “COMMON” is included in the uploaded annotation file, it is replaced with information from “1. Contact person”, “2. Hold date”, “3. Submitter”, and “4. Reference.”
- No need to prepare the COMMON block. The values entered in each form of “1.Contact person” - “4.Reference” are used for the actual submission even if you have included the COMMON to the annotation file.
- You should include the biological feature to the annotation file.
- Related page
- Definition of Feature key / Definition of Qualifier key / Organism qualifier / Protein Coding Sequence; CDS feature
Common mistake that causes file-uploading error
-
When you use Excel for making annotation file, you need to copy it to text editor and save as the text file. The annotation file must be saved as tab-deliminated text file format.
-
Entry names of the nucleotide sequences entered at “5. Sequence” are not described in the annotation file, or order of each entry names is different between the annotation file and the nucleotide sequence.
-
The tab column structure is wrong.
-
Some extra space or illegal characters, such as multibyte, unicode, or unprintable character, are included in the file.
-
The annotation contains only source feature. Add appropriate featute(s) to the annotation.
Error/Warning
-
If there are is no error after you click “Confirm”, “Next” button is changed to be clickable and you can move to the next process.
-
Error/warning messages are displayed beneath annotation input area when error or warning occurred.
-
In order to correct error, please scroll-up the screen, and edit entry at which error occurred on annotation input field. Please click “Confirm” after you correct the errors.
-
When error/warning occurred at “Submitter”, “Reference”, or “Sequence”, please go back to previous page by clicking the page name on progress bar. After correction, you must click “Next” on each page, and then click “7.Annotation” on progress bar in order to return to “7. Annotation” page.
-
“Next” button changes to be clickable, even though there are some warnings. Please check again the input data. You should correct if there are any problems. You can click “Next” if you believe that there is no problem in the input data.
For detailed error/warning messages, please refer “validator error message”. Add “#JPxxxx” (xxxx = 4 digits) after the URL for direct link to each page.
e.g. http://www.ddbj.nig.ac.jp/ddbj/validator-e.html#JP0015 … Please be patient until the page scrolls to the corresponding error number.
How to obtain amino acid sequence
Please refer to the page, “How to confirm translated amino acid sequences (i.e. /translation qualifier) for CDS features”.
As an alternative way, you can use the following Web services.
Final page
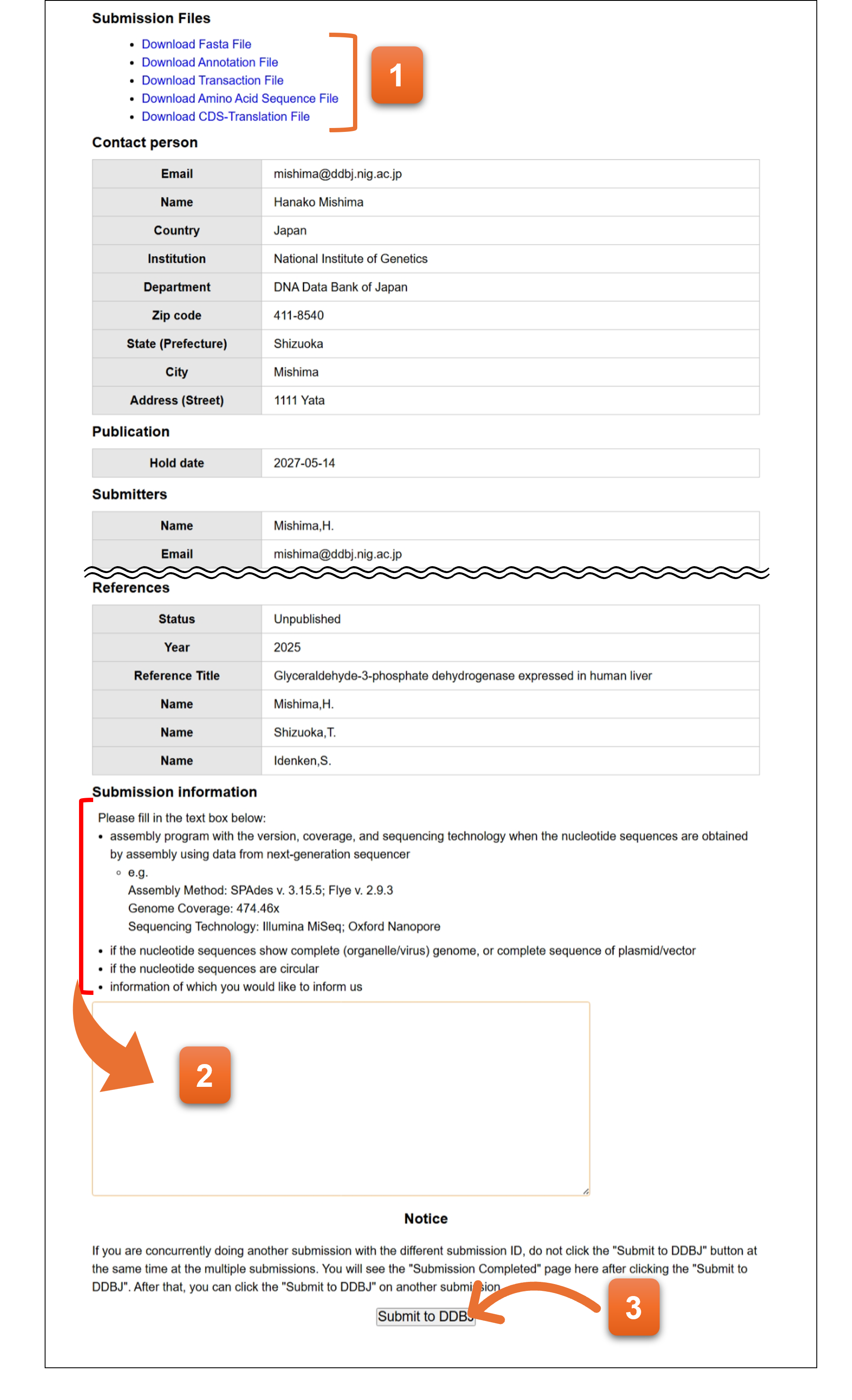
Final confirmation screen. Please enter the additional information when applicable.
- You can download the files in the format of DDBJ annotation file and nucleotide sequence file.
- Please describe additional information to the text area when the submission corresponds to the listed items.
- Please click to complete the submission. You will not be able to go back to the previous page after clicking the button.
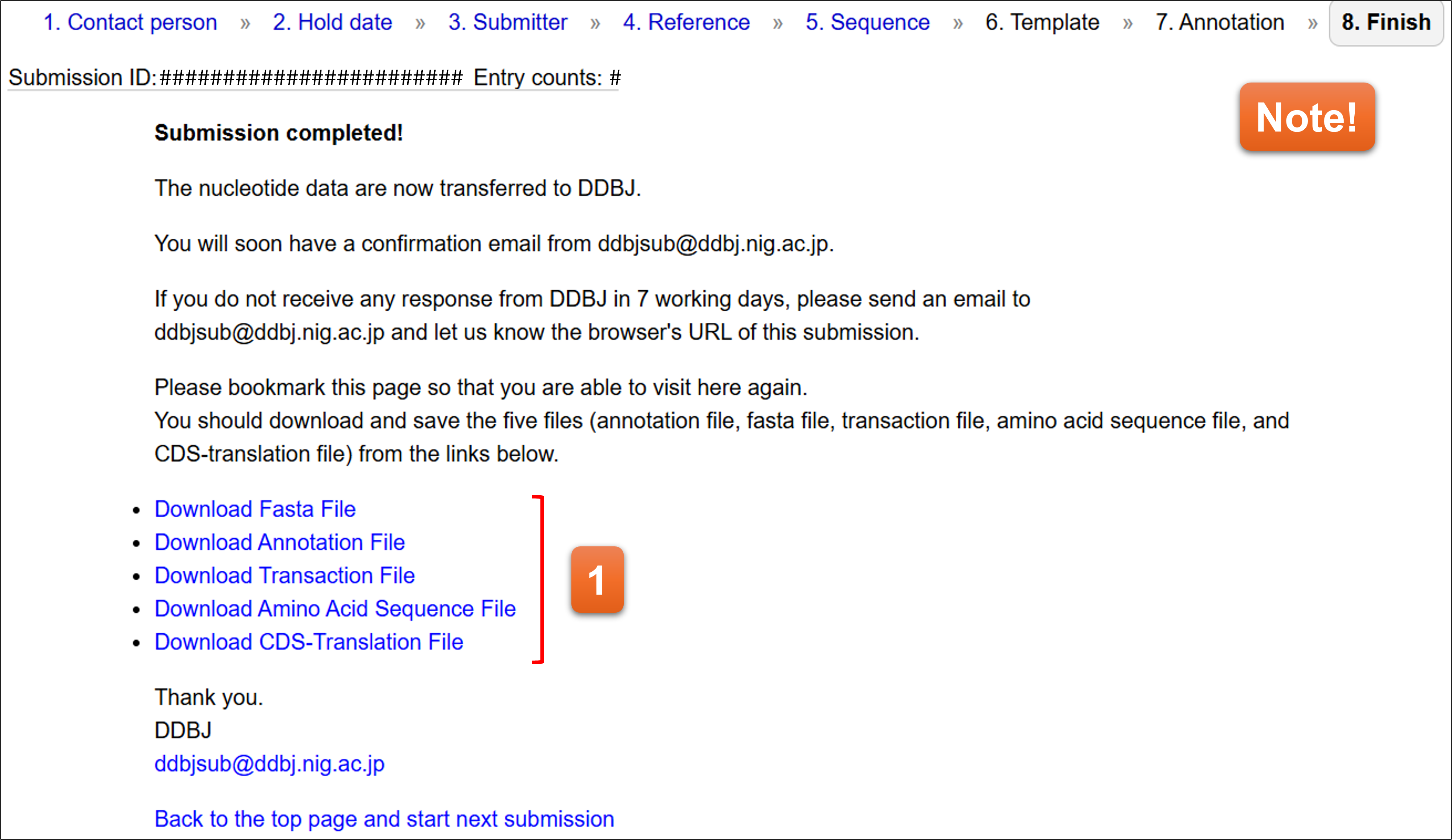
8. Finish
You have completed the submission when you see the screen. The data are automatically transferred to DDBJ registration server, and an email is sent to contact person’s email address.
- You can download the files in the format of DDBJ annotation file and nucleotide sequence file. These files will be transferred to DDBJ server for the registration.
📌Note! Please bookmark the page. You can access only “Finish” page for a certain period of time after finishing the submission.
Email to notify the completion